Sir,
Cowpox is a rare, zoonotic infection caused by an orthopoxvirus. Despite the name, the infection is rarely transmitted from cattle to humans. Since 1985 there have been several reports of cowpox transmitted to humans from domestic cats, which are considered the most common source of human infection (1–4). The cats acquire the infection from small wild rodents, which are regarded as the principal reservoir. The infection is usually transferred to humans through lesions on exposed skin, and most often the lesion is localized on the hands or in the face, at the site of primary inoculation. Infection via the mucosa has also been reported, presenting as facial cellulitis caused by infection of the respiratory epithelium of the nose (5).
More than 70% of infected individuals have a solitary lesion (6). Multiple lesions may result from multiple primary inoculation sites, autoinoculation or lymphatic spread with a sporotricoid distribution (7). A preponderance of males aged 12–18 years with facial lesions has been reported (6). In the majority of cases the course is mild and self-limiting, but severe generalized infections have been described in patients with atopy or immunosuppression.
CASE REPORT
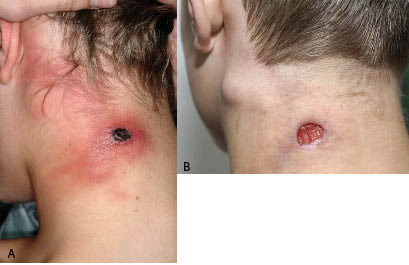
A 13-year-old boy living in a rural environment was referred to the Department of Dermatology at Odense University Hospital with an ulcerating skin lesion on the left side of his neck, accompanied by a pronounced local lymphadenopathy (Fig. 1). The patient lived on a farm and had close contact with cats, dogs and sheep, but there was no history of symptoms in these animals. Two weeks before referral, he developed a small papule on the left side of the neck, which gradually enlarged and developed into a pustule. He initially had systemic symptoms with high-grade fever up to 39.5ºC and flu-like symptoms. At this stage he sought medical attention by an otologist who initially prescribed dicloxacillin and, when this had no effect, erythromycin, as bacterial infection was suspected. The fever abated in 4–5 days, but the neck lesion continued to enlarge and ulcerated during the second week. At the time of referral the patient presented with an indurated, indolent ulcer, 3 cm in diameter, with a hard, black eschar surrounded by erythema and local lymphadenopathy. He was treated with oral antibiotics until the viral aetiology was established and local antiseptic treatment was continued to prevent secondary infection. The lymphadenopathy gradually subsided and the ulcer healed in 6–8 weeks with some scarring.
Fig. 1. (A) Skin lesion in the left side of the neck with a black eschar surrounded by erythema. (B) Pronounced lymphadenopathy after 4 weeks.
Laboratory investigations revealed a slightly raised erythrocyte sedimentation rate, while leukocyte counts, C-reactive protein and liver function tests were normal. Cutaneous anthrax and impetigo were excluded by negative swab culture from the ulcer. Swabs from the lesion were negative, as were serological tests for Bartonella henselae and Francisella tularensensis. Biopsy samples obtained for Mycobacterium tuberculosis culture were also negative.
Two 4-mm punch biopsies were taken from the rim of the ulcer. Light microscopy of haematoxylin and eosin (H&E)-stained sections showed non-specific inflammatory changes in the epidermis and dermis. No cytoplasmic inclusions were seen.
The swab sample was analysed initially using MagNA Pure extraction method (Total NA Isolation Kit, no. 3 038 505, Roche, Mannheim, Germany). To improve sensitivity, an additional extraction was performed subsequently, using increased sample volumes (1000 µl) combined with decreased elution volumes (50 µl). Ulcer biopsy material was split and prepared for electron microscopy and cultivation of mycobacteria, for which the latter was treated with BLL™ MycoPrep™ (BD Diagnostics, Franklin Lakes, NJ, USA) according to the manufacturer’s instructions. This material was extracted using the modified protocol.
Three polymerase chain reaction (PCR) assays were performed on extracted material. Initially, a real-time PCR assay targeting the F-gene was performed (8). To confirm the findings and to perform genotypic analysis, diagnostic PCR assays were performed amplifying regions of approximately 940 and 387 bp within the haemagglutinin (HA) and the A13L genes, respectively.
The A13L assay was performed essentially as described elsewhere, using the Platinum kit (cat. no. 10966-018, Invitrogen, Carlsbad, CA, USA) (9). The HA-specific assay was modified from Ropp et al. (10) including an additional set of PCR primers specific for monkeypox virus (MoPoxF: 5’-atgacacaattaccaatac-3’, MoPoxR: 5’-ctagactttgttctctg-3’). In brief, using the Platinum kit, the final concentration of all 4 primers was 200 nM, in 4 mM MgCl2. After initial hot-start (94ºC 2 min), amplification included 40 PCR cycles of 30 s at 94ºC, 15 s at 51ºC, and 60 s at 72ºC, and a final 5-min elongation at 72ºC. PCR reactions were analysed by agarose gel electrophoresis.
Specific amplicons from the HA and A13L PCR assays were purified using QIAquick PCR product purification kit (Qiagen, Germany). HA and A13L PCR products were sequenced using the PCR primers in standard sequencing reactions according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA, USA) and sequences submitted to GenBank (DQ675140 and DQ675141, respectively). Sequences between the primer-binding regions were subjected to homology search against available GenBank sequences. In addition, phylogenetic analysis were performed including isolates of camelpox (CMLV), cowpox (CPXV), ectromelia (ECTV), monkeypox (MPXV), taterapox (GBLV), vaccinia (VACV), buffalopox (BPXV), rabbitpox (RPXV) and variola (VARV). From a Clustal W nucleotide sequence alignment, neighbour-joining phylogenic trees were constructed using MEGA 3.1 software (11). The abbreviations from International Committee on Taxonomy of Viruses, as well as GenBank accession numbers, are included for every sequence.
Initially only the F-gene specific real-time PCR assay was positive. Melting point analysis clearly indicated that the PCR product derived from a non-variola orthopox species.
To improve sensitivity and thereby enable confirmation and genotyping via the HA and A13L assays, the extraction procedure was modified. The second extract of the swab was positive in both the F- and HA-specific PCR assays, whereas the A13L was not. The extracted material from the biopsy was positive in all 3 orthopox-specific PCR assays. We found no indications of PCR inhibition. Thus, for PCR using an improved method of extraction and MycoPrep™-treated biopsy material increased sensitivity and allowed for genotypic analysis.
Purified amplicons from the HA and A13L PCR assays were sequenced in both directions. By BLAST (Basic Local Alignment Search Tool) homology searches, both the HA and A13L sequences showed the greatest homology to a number of cowpox isolates, thereby confirming the diagnosis. The sequences of the HA and A13L amplicon have been submitted to GenBank (DQ675140 and DQ675141, respectively).
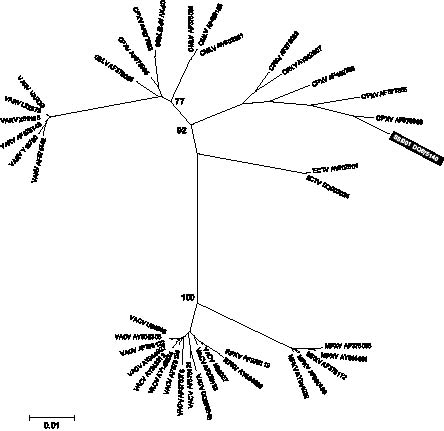
All available non-identical orthopox database sequences from the HA and A13L regions were aligned using the neighbour-joining algorithm with default Kimura 2-parameter settings (Fig. 2). For HA the sequences obtained from the patient samples cluster together with other cowpox isolates. Thus, the alignment substantiates the homology search. Electromyography (EM) revealed no orthopoxvirus.
Fig. 2. Phylogenetic relationship between the patient virus and available reference haemagglutinin sequences. The tree was constructed using the neighbour-joining algorithm, with default Kimura 2-parameter model settings in MEGA 3.1 software (11). The phylogenetic distance scale bar indicates estimated changes per nucleotide. Bootstrap (1000 replicates) values greater than 70 are indicated on the major tree nodes.
DISCUSSION
In recent decades sporadic cases of cowpox have been reported in European countries, including Denmark. The present finding and one earlier case-report (12) clearly demonstrate that cowpox infections occur in Denmark, although they are probably rare. In comparison human cowpox cases have been reported regularly in England and Germany (2–4, 6).The geographic distribution of cowpox virus extends to most neighbouring European countries (13, 14).
The boy in the present case probably acquired the infection from contact with his cats, but further serological investigations are being undertaken to ascertain a possible transmission route.
A survey of Danish cats revealed that approximately 5% are seropositive, comprising both farm cats and town cats, which is low in comparison with neighbouring countries (L.S. Christensen, personal communication).
The diagnosis of cowpox is based on its characteristic clinical appearance and is to be suspected if there is a history of contact with cats or rodents. The most important differential diagnoses include cutaneous anthrax, herpes, parapoxvirus infections and impetigo. The allocation of resources to laboratory techniques for detection of agents related to potential bioterrorism has improved the possibilities for fast and reliable identification of poxvirus infections. Thus, we have 3 PCR assays confirming the cowpox finding. We used 3 different extractions from 2 different sample materials. All assays confirmed ortho/cowpox infection. The sensitivity of the PCR assays is approximately 6 copies/sample for the F-gene (14), whereas the HA and A13L has a lower detection level, around 100 copies/sample. Thus sensitive PCR enables rapid diagnosis without preceding virus cultivation, as reported elsewhere (4). The inability to detect the cowpox infection with EM analysis of the biopsy material may be explained by the sample material and the sampling time. In other studies virus particles were detectable by EM from ulcer (1–3, 12) and/or after cultivation (1, 2, 4).
The 3 primer sets used are specific for orthopox virus only (8–10). This fact, combined with the results of the homology, excludes the possibility of infection by one of the parapox virus types known to infect humans; pseudocowpox virus (causing milker’s nodule) or bovine papular stomatitis virus.
As in earlier reports we used sequence analysis to establish the genotype of the virus (12), whereas other studies have relied on analysis of restriction enzyme-digested PCR amplicons (4, 5). In our laboratory setting, sequence analysis is performed on a routine basis and requires no type-specific positive control material. Restriction enzyme digest analysis provides no additional information as compared to complete determination of the DNA sequence.
The HA-specific PCR amplifies > 900 bp, which is more than is obtained from the A13L amplicon (390 bp). In addition, there are markedly fewer full-length A13L reference sequences published compared with HA. Together, this makes the amplified HA region suitable for phylogenetic analysis.
It has been suggested that variola and monkeypox virus have evolved from cowpox-like virus (15). This may well explain the greater sequence variation among cowpox isolates compared with variola and monkeypox, as reported earlier (15). Expectedly, in our phylogenetic analysis the cowpox reference sequences fall in more than one cluster.
Vaccination against smallpox, which also confers protection against cowpox infection, was discontinued in Denmark in 1976. After 3 decades it is estimated that more than one-third of the Danish population is susceptible to infection by cowpox as well as other orthopoxvirus. Considering the number of susceptible individuals we speculate that a number of cowpox infections in Denmark are not diagnosed. A survey of seroprevalence is needed to establish this.
REFERENCES
1. Heilbronner C, Harzic M, Ferchal F, Pothier A, Charara O, Beal G, et al. Cowpox virus infection in a child. Arch Pediatr 2004; 11: 335–339.
2. Hawranek T, Tritscher M, Muss WH, Jecel J, Nowotny N, Kolodziejek J, et al. Feline orthopoxvirus infection transmitted from cat to human. J Am Acad Dermatol 2003; 49: 513–518.
3. Haenssle HA, Kiessling J, Kempf VA, Fuchs T, Neumann C, Emmert S. Orthopoxvirus infection transmitted by a domestic cat. J Am Acad Dermatol 2006; 54: S1–S4.
4. Schupp P, Pfeffer M, Meyer H, Burck G, Kolmel K, Neumann C. Cowpox virus in a 12-year-old boy: rapid identification by an orthopoxvirus-specific polymerase chain reaction. Br J Dermatol 2001; 145: 146–150.
5. Pahlitzsch R, Hammarin AL, Widell A. A case of facial cellulitis and necrotizing lymphadenitis due to cowpox virus infection. Clin Infect Dis 2006; 43: 737–742.
6. Baxby D, Bennett M, Getty B. Human cowpox 1969–93: a review based on 54 cases. Br J Dermatol 1994; 131: 598–607.
7. Motley RJ, Holt PJ. Cowpox presenting with sporotricoid spread: a case report. Br J Dermatol. 1990; 122: 705–708.
8. Olson VA, Laue T, Laker MT, Babkin IV, Drosten C, Shchelkunov SN, et al. Real-time PCR system for detection of orthopoxviruses and simultaneous identification of smallpox virus. J Clin Microbiol 2004; 42: 1940–1946.
9. Pulford DJ, Meyer H, Ulaeto D. Orthologs of the vaccinia A13L and A36R virion membrane protein genes display diversity in species of the genus Orthopoxvirus. Arch Virol 2002; 147: 995–1015.
10. Ropp SL, Jin Q, Knight JC, Massung RF, Esposito JJ. PCR strategy for identification and differentiation of small pox and other orthopoxviruses. J Clin Microbiol 1995; 33: 2069–2076.
11. Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004; 5: 150–163.
12. Christensen LS, Nielsen EB, Nowicki J, Andersen J, de Stricker K. Påvisning af kokoppevirus (cowpoxvirus) i Danmark. [Detection of cowpox virus in Denmark]. Ugeskr Laeger 2005; 167: 1646–1647 (in Danish).
13. Chantrey J, Meyer H, Baxby D, Begon M, Bown KJ, Hazel SM, et al. Cowpox: reservoir hosts and geographic range. Epidemiol Infect 1999; 122: 455–460.
14. Baxby D, Bennett M. Cowpox: a re-evaluation of the risks of human cowpox based on new epidemiological information. Arch Virol Suppl 1997; 13: 1–12.
15. Esposito JJ, Fenner F. Poxviruses. In: Fields BN, Howley PM, Knipe DM, editors. Fields virology 3rd edn. Philadelphia: Raven Publishers, 1996: p. 2885–2921.