Hiraku Suga1, Yuichiro Tsunemi1, Makoto Sugaya1, Satoru Shinkuma2, Masashi Akiyama2, Hiroshi Shimizu2 and Shinichi Sato1
Departments of Dermatology, 1Faculty of Medicine, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8655, and 2Hokkaido University Graduate School of Medicine, Sapporo, Japan. E-mail: hiraku_s2002@yahoo.co.jp
Accepted December 16, 2010.
Localized autosomal recessive hypotrichosis (LAH) 2 is a type of non-syndromic human alopecia that is inherited as an autosomal recessive trait. We describe here a patient with LAH2 who had mutations in the lipase H (LIPH) gene. We analysed hair shaft morphology using light and scanning electron microscopy (SEM). In addition, we review the features of other non-syndromic human alopecias.
CASE REPORT
The patient was a 4-year-old boy, the firstborn of healthy and unrelated Japanese parents, born after an uneventful pregnancy. He had scant hair at birth, which grew very slowly in infancy.
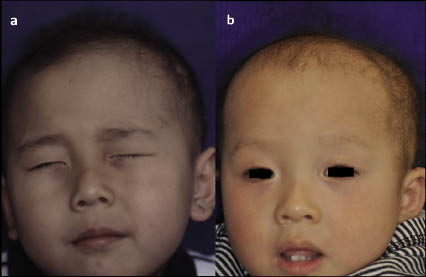
Clinical examination revealed hypotrichosis of the scalp (Fig. 1a). The hairs were sparse, thin, and curly, and not easily plucked. The left eyebrow hair was sparse, but the eyelashes and other body hair were present in normal amounts. Teeth, nails, and the ability to sweat were completely normal. Clinical features of keratosis pilaris, milia, scarring, and palmoplantar keratoderma were absent. Psychomotor development was normal. The patient’s younger brother also had severe hypotrichosis; since birth his hair was curly, and his eyebrow hair virtually absent (Fig. 1b). No other family members, including his parents, had similar hair abnormalities. Laboratory tests of the patient showed normal serum levels of copper and zinc, and liver and kidney function tests were all within normal ranges. Over a period of 2 years there was no improvement or exacerbation of hypotrichosis in the patient.
Fig. 1. (a) Clinical features of the patient at 4 years of age. (b) Clinical features of the younger brother at 1 year 4 months of age. Permission is given from the parents to publish these photos.
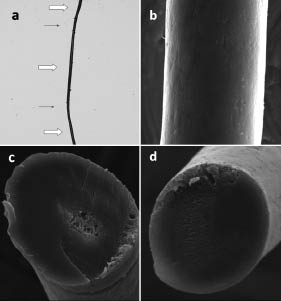
Light microscopy of the patient’s scalp hairs revealed that approximately 10% had structural abnormalities. Abnormal hairs were composed of thick dark parts and thin light parts (Fig. 2a). SEM revealed alterations of the cuticular architecture. Cuticular cells were absent from both the thick and thin parts (Fig. 2b). Cross-sectional observation showed that thick, but not thin, sections had hair medulla (Fig. 2c, d). Light microscopy on hairs from the patient’s younger brother revealed that they were composed of thin and thick parts (data not shown).
Based on the clinical features, hair microscopy and family pedigree, we suspected LAH2 or LAH3. To determine the type of LAH, we looked for gene mutations in LIPH and LPAR6 (encoding lysophosphatidic acid receptor 6). Two prevalent missense mutations in LIPH were found (1); c.736T>A (p.Cys246Ser) and c.742C>A (p.His248Asn). The mutations were carried in a compound heterozygous state. No mutations were found in LPAR6. The parents did not consent to genetic testing of the younger brother or themselves.
Fig. 2. (a) Light microscopy (×40). Hair was composed of thick () and thin parts (→). (b) Scanning electron microscopy (×900). Cuticular cells were absent in both thick and thin sections. (c, d) Scanning electron microscopy (cross-section, ×900). (c) Thick regions showed hair medulla, while (d) thin regions did not.
DISCUSSION
The different LAH subtypes map to chromosomes 18q12.1, 3q27.3 and 13q14.11–13q21.32, and are designated LAH1, LAH2 and LAH3, respectively (2–4). Mutations in DSG4 (encoding desmoglein 4) have been found to be responsible for LAH1 (5). Kazantseva et al. (6) reported deletion mutations in LIPH leading to LAH2. Pasternack et al. (7) reported disruption of LPAR6 in families affected with LAH3.
Table I summarizes of genetic, non-syndromic human alopecias. In hypotrichosis simplex of the scalp, hair loss is limited to the scalp without hair shaft abnormalities. The causative gene is CDSN (encoding corneodesmosin) on 6p21.3 (8). The clinical presentations of monilethrix vary among patients. Mild cases have hair loss limited to the scalp, while severe cases show generalized alopecia. Hair shaft abnormalities are characteristic, demonstrating regularly-spaced, spindle-shaped swellings. The nodes are as thick as normal hair and the atrophic internodes represent areas where the hair is easily broken. Causative genes are hHb1, hHb3 and hHb6 (12q13) (9), which encode for basic hair keratins.
Table I. Features of genetic, non-syndromic human alopecias
|
Disease (ref)
|
Hair shaft shape
|
Eyelash/eyebrow
|
Causative gene
|
Mode of inheritance
|
|
Hypotrichosis simplex of scalp (8)
|
Normal
|
Normal
|
CDSN
|
Autosomal dominant
|
|
Monilethrix (9)
|
Regularly spaced, spindle-shaped swellings
|
Absent to normal
|
hHb1, 3, 6
|
Autosomal dominant
|
|
Atrichia with papular lesions (10)
|
Normal
|
Absent
|
HR
|
Autosomal recessive
|
|
Hypotrichosis, Marie Unna type (11)
|
Iron-wire
|
Sparse
|
U2HR
|
Autosomal dominant
|
|
Hereditary hypotrichosis simplex (12)
|
Short, thin, easily plucked
|
Absent to sparse
|
APCDD1
|
Autosomal dominant
|
|
Localized hereditary hypotrichosis (LAH1) (2, 5, 13)
|
Moniliform
|
Absent to normal
Absent to normal
Absent to normal
|
DSG4
|
Autosomal recessive
|
|
Localized hereditary hypotrichosis (LAH2) (3, 6, 14)
|
Curled
Curled
|
LIPH
|
Autosomal recessive
|
|
Localized hereditary hypotrichosis (LAH3) (4, 7, 15)
|
LPAR6
|
Autosomal recessive
|
In case of atrichia with papular lesions, hair loss on the entire body occurs several months after birth. The gene responsible is HR (encoding ”hairless”) (10), a transcription modulating factor that influences the regression phase of the hair shaft cycle. Patients with hypotrichosis, Marie Unna type have hard and rough scalp hair, described as iron-wire hair. Generalized hypotrichosis is often seen. U2HR, an inhibitory upstream open reading frame of the human hairless gene (11), is mutated in this condition. Hereditary hypotrichosis simplex is characterized by hair follicle miniaturization. The defective gene is APCDD1 (encoding adenomatosis polyposis down-regulated 1) (12). Hairs are short, thin, and easily plucked. Eyelashes and eyebrows are also affected.
As already mentioned, there are three types of localized hereditary hypotrichosis. LAH1 patients have hair shaft abnormalities that resemble moniliform hair (13). LAH1 can be viewed as an autosomal recessive form of monilethrix. Patients with LAH2 and LAH3 have woolly hair (14, 15), and eyelashes and eyebrows are often sparse or absent. Upper and lower limb hairs are sometimes absent too.
Our patient had hypotrichosis of the scalp with sparse left eyebrow hair and irregularly spaced segments of thick and thin hair, but not to a degree that could be labelled moniliform. The mode of inheritance was autosomal recessive and LIPH was found to be abnormal, thus establishing a diagnosis of LAH2. One of the mutations (c.736T>A) leads to an amino acid change (p.Cys246Ser) of a conserved cysteine residue, which forms intramolecular disulphide bonds in the lid domain in the structure model of LIPH (1). The other mutation (c.742C>A) results in alteration of one of the amino acids of the catalytic triad (Ser154, Asp178, and His248) of LIPH (p.His248Asn) (1).
Regarding hair shaft morphology, Horev et al. (14) reported that hairs of LAH2 patients showed decreased diameter under light microscopy. This is the first report to describe hairs from an LAH2 patient by SEM. Shimomura et al. (13) observed hairs of LAH1 patients by SEM and found variable thickness of the hair shaft, resulting in nodes and internodes. Which are absent in LAH1 (our observation). Longitudinal ridges and flutes were observed at internodes, and the breaks always occurred at internodes in LAH1. These features resemble those of moniliform hair rather than LAH2. However, in the end gene analysis is probably easier to accomplish than SEM to distinguish the two types of LAH.
ACNOWLEDGEMENTS
We thank Dr Andrew Blauvelt, Department of Dermatology, Oregon Health & Science University, for many helpful comments.
REFERENCES