Koen D. Quint1, Jasper I. van der Rhee1, Nelleke A. Gruis1,2, Jeanet A. ter Huurne2, Ron Wolterbeek3, Nienke van der Stoep2, Wilma Bergman1 and Nicole A. Kukutsch1
1Department of Dermatology, 2Centre for Human and Clinical Genetics and 3Department of Medical Statistics, Leiden University Medical Centre, Leiden, The Netherlands
Individuals with two red hair colour (RHC)-MC1R genetic variants have light skin and blond /reddish hair and, in comparison with those without such alleles, are at an increased risk of developing melanoma. Our study investigated the association of RHC variants and the Total Dermoscopy Score (TDS), and the items that make up the TDS, in those with atypical naevi and melanomas from high risk melanoma patients. Eight hundred and seventy-six atypical naevi and 21 melanomas were scored according to the TDS system and MC1R polymorphisms were determined. Analyses revealed that several TDS items including pigment network, dark brown colour and streaks were more frequently
observed in atypical naevi from individuals without RHC variants, while structureless areas were more often observed in individuals with 2 RHC variants. Finally, no significant difference in TDS was detected in atypical naevi from individuals with two RHC variants compared to those without RHC. Clinicians should be aware of a different dermoscopic naevus phenotype in patients with light blond or RHC MC1R variants. Key words: CDKN2A; dermoscopy; MC1R;
atypical naevi; melanoma; red hair; total dermoscopy score.
(Accepted August 13, 2012.)
Acta Derm Venereol 2012; 92: XX–XX
Wilma Bergman, MD, PhD, Department of Dermatology, LUMC Leiden, Albinusdreef 2, NL-2300 RC Leiden, The Netherlands. E-mail: w.bergman@lumc.nl
In the past decade, the incidence and mortality rates for cutaneous malignant melanoma in (CMM) Caucasions have increased (1, 2). In the majority of individuals in the general population, activation at the Melanocortin 1 Receptor (MC1R) leads to an increase in the ratio of eumelanin to pheomelanin: eumelanin (black/brown colour) is UV protective, relative to the potentially carcinogenic pheomelanin (red/yellow colour) (3). Several polymorphisms of the MC1R cause an increase in the pheomelanin/eumelanin ratio, favoring the carcinogenic pheomelanin. These polymorphisms of MC1R are clinically associated with red hair colour (RHC), fair skin, poor tanning ability and a higher risk for development of CMM (4–8). Besides the well established association between the MC1R variants D84E, R151C, R160W and D294H and RHC, the less frequent MC1R polymorphisms I155T and R160Q also show an association with RHC (9, 10). These MC1R polymorphisms are considered as low penetrance genes, since those polymorphisms are relatively common compared to the incidence of CMM.
A substantial risk factor for the development of CMM is a genetic predisposition in high penetrance genes. Twenty-five to 50% of individuals who have had CMM and a strong family history (with one first degree or two second degree family members with CMM) carry mutations in high penetrance genes such as the Cyclin Dependent Kinase Inhibitor 2A (CDKN2A) gene and the Cyclin Dependent Kinase 4 (CDK4) gene (11–13). At this moment, the CDKN2A gene is the most established high penetrance gene, encoding for the two tumour suppressor proteins p16 and p14ARF (14). A founder mutation, consisting of a 19bp deletion in exon 2 of the CDKN2A gene, was detected in Dutch families originally living in the Leiden area and is therefore known as the p16-Leiden founder mutation (15).
Individuals with a genetic predisposition, are strongly recommended to undergo periodic screening of the total body skin. In this population, dermatologists are tasked with detecting minor abnormalities in melanocytic lesions as early as possible. Dermoscopy can form a bridge between the clinical diagnosis and the histopathological diagnosis, by revealing morphological structures and colours not visible to the naked eye and improve the sensitivity and specificity of the clinical CMM diagnosis (16–18).
A recent study suggested that the diagnosis of early melanoma using the ABCD Total Dermoscopy Score (TDS) is more difficult in CDKN2A mutation carriers with two MC1R RHC variants than in CDKN2A mutation carriers with 0 MC1R RHC variants (19). The authors suggested that an integrated approach including medical history, clinical investigation and dermoscopic data would be useful in those high-risk patients.
Atypical naevi (AN) are considered precursor and indicator lesions of melanoma. Therefore, it is important for the clinician to know whether the dermoscopic features in AN and melanomas from carriers of 2 RHC variants differ from those with 0 RHC variants (20). The objective of the current study was to explore the relation between RHC variants and dermoscopic items (as scored by the ABCD-rule of dermoscopy) in AN and melanoma among CDKN2A founder mutation carriers.
MATERIALS AND METHODS
Patient and atypical naevi selection
Between January 2000 and July 2002, 519 members from 18 proven CDKN2A mutated families were invited to have dermoscopic images taken of their AN with the MoleMax II™ (Derma Medical Systems, Vienna, Austria). Participants were invited based on the clinical diagnosis of familial melanoma, independently of the CDKN2A mutation status and naevus phenotype. They were examined by a dermatologist, who marked all melanocytic naevi for recording. Inclusion criteria for lesions were: (i) at least partly a flat component, (ii) a minimal diameter of 5 mm in at least one direction, and (iii) at least two of the following criteria: hazy border, colour variegation, or a red hue. Subsequenlty a research nurse recorded baseline dermoscopic images of all marked lesions and took macroscopic body mapping images. Patients’ history of melanoma was recorded and blood samples were taken to determine the CDKN2A mutation status of each individual patient as previously described (7). All lesions suspected to be possible melanomas were excised within two weeks after recording of the naevi. AN located on the head, feet and hands were excluded from dermoscopic analysis, since the ABCD-pattern is not suitable for those lesions. In total, 111 proven CDKN2A mutation carriers with 876 AN were included for dermoscopic analysis and MC1R sequencing (Fig. 1). Inclusion was based on the availability of both dermoscopic images in the MoleMax™ and blood.
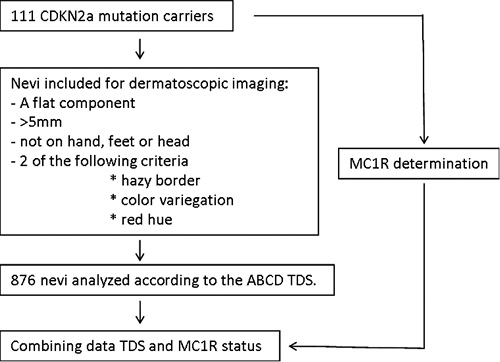
Fig. 1. A flowchart summarizing the method of patient selection, naevi selection and MC1R testing.
Melanomas selection
All available digital images from melanomas of the 111 CDKN2A mutation carriers, diagnosed between 2001 and 2009 were selected. Pathology reports summarising the stage of the melanoma were collected. In total 21 melanomas from 17 CDKN2A mutation carriers were included for dermoscopic analysis.
Dermoscopic analysis
The melanocytic lesions were scored according to the ABCD rule of dermoscopy which is a semiquantitive numerical approach (21). The ABCD rule calculates a Total Dermascopy Score (TDS) based on the asymmetry (A), border (B), colour (C) and differential structures (D). In evaluating the asymmetry, the lesions were bisected by two 90° axes that were positioned to produce the lowest possible asymmetry score. A value between 0 (no asymmetry) and 2 (asymmetry on both axes) was scored for asymmetry. The evaluation of the border score is determined by a sharp abrupt cut-off of the pigment pattern. The naevi were divided into 8 parts, making the minimum score 0 (no parts showed an abrupt cut-off) and the maximum score 8 (all parts showed an abrupt cut-off). The evaluation of the colours can lead to a maximum score of 6 (white, red, light brown, dark brown, blue-gray and black). The differential structures had a maximum score of 5 (pigment network, dots (min 3), globules (min 2), streaks (min 3) and a structureless area > 10% of the lesion). To generate a TDS, the individual scores of ABCD were multiplied by the coefficients 1.3 for asymmetry, 0.1 for border, 0.5 for colours and 0.5 for structures. A lesion with a TDS < 4.75 was suggestive of a benign melanocytic lesion, a lesion with a TDS between 4.75 and 5.45 was suggestive of a dysplastic naevus and a lesion with a TDS > 5.45 was suggestive of a melanoma. All lesions were scored in consensus by a trained resident (KQ) from the dermatology department and a dermatologist, specialized in melanocytic lesions and dermoscopy (NK). Both were aware of the CDKN2A outcome, but were not aware of MC1R outcome and the patient’s phenotype.
MC1R sequencing
The MC1R gene consists of a single coding exon of 954 bp. For sequencing, the gene is divided into three covering PCR fragments; 1A, 1B and 1C. Fragment 1A starts at 5’UTR c.-73 until c.377, fragment 1B covers c.356 to c.717 and fragment 1C c.713 until c*16 at the 3’UTR. A total of 3 forward and 3 reverse primers were used. The primer sequences were respectively: MC1Rex1AF1;TGTAAAACGACGGCCAGTGCAGCACCATGAACTAAGCA, MC1Rex1AR1; CAGGAAACAGCTATGACCAGGCTGGACAGCATGGAG, MC1Rex1BF1;TGTAAAACGACGGCCAGTTGCAGCAGCTGGACAATG, MC1Rex1BR1; CAGGAAACAGCTATGACCAGGATGGTGAGGGTGACAGC, MC1Rex1CF1; TGTAAAACGACGGCCAGTACCAGGGCTTTGGCCTTA and MC1Rex1CR1; CAGGAAACAGCTATGACCCTCTGCCCAGCACACTTAAA.
Amplification of the 3 fragments were performed in 25 μl PCR reactions containing 150 ng of genomic DNA, 5x Colourless GoTaq Reaction buffer containing 7.5 mM MgCl2 (Promega, Madison, WI, USA), 6 pmol of each primer, 200 μM of each deoxynucleotide triphosphates (dNTP) and 1 U of Taq DNA polymerase (Promega). Fragment 1A reaction mix contained an extra 10% dimethyl sulphoxide (DMSO) and 1x PCRx Enhancer Solution (Invitrogen, Life Technologies, Grand Island, NY, USA). Cycle conditions are the same for fragment 1B and 1C, starting with a hot start at 95°C, initial denaturation for 5 min at 95°C, followed by 35 cycles of 30 s 94°C denaturation, 30 s at 55°C annealing and 30 s at 72°C elongation, concluding with a 5 min extension at 72°C. Fragment 1 A only differs in the annealing temp (60°C).
The PCR products were first purified with the AMPure PCR Purification kit (Beckman Coulter, Indianapolis, USA) before sequencing.The MC1R gene was sequenced by using the standards sequencing kit of Applied Biosystems (3730 DNA Analyzer Sequencing Standards, BigDye® Terminator v3.1) and submitted at an ABI PRISMTM 3730 DNA Analyzer. Sequence data were analysed utilizing the program SeqScape Software version 2.5 (Applied Biosystems).
Statistical analysis
Statistical analyses were performed using the Statistical Package for the Social Science (SPSS version 17.0; Gorinchem, the Netherlands). The TDS results were crudely analyzed with a Student’s t-test. A Levene’s test was used to determine the equality of the variances. Secondly, the TDS was analyzed using a Linear Mixed Model. Taking into account the varying numbers of AN within patients, the patients were included in the model as a random effect and RHC as a fixed effect. The crude analyses of the differential structures and colours of the AN were performed using Pearson’s chi-squared test. Asymmetry, Border, Colours and Differential structures were all binary outcome variables and RHC was used as predictor. Again, a second analysis, repeated measures logistic regression (Generalized Estimating Equations) was performed, taking account of the repeated measurement of the outcome variables (multiple AN per patient). A p-value ≤ 0.05 was considered significant.
RESULTS
Distribution of MC1R variants among patients with atypical naevi
A total of 98 MC1R polymorphisms were determined among the 111 CDKN2A mutation carriers and 49 MC1R polymorphisms (50%) were determined as RHC variants (Table I). Among the CDKN2A mutation carriers, 72 individuals (64.9%) had no RHC variants, 29 individuals (26.1%) had 1 RHC variant and 10 individuals (9.0%) had 2 RHC variants. In total, 513 AN (58.6%) were observed in individuals with 0 RHC variants, 253 AN (28.9%) were observed in individuals with one RHC variant and 110 AN (12.6%) were observed in individuals with 2 RHC variants.
Table I. Frequency of MC1R variants among 111 CDKN2A mutation carriers
|
Polymorphism |
Variant |
n (%) |
|
V60L |
nRHC |
19 (19.4) |
|
V92M |
nRHC |
19 (19.4) |
|
R151C |
RHC |
20 (20.4) |
|
I155T |
RHC |
4 (4.1) |
|
R160W |
RHC |
22 (22.4) |
|
R160Q |
RHC |
3 (3.1) |
|
R163Q |
nRHC |
11 (11.2) |
|
Total |
|
98 (100) |
RHC: red hair colour; nRHC: no red hair colour.
Dermoscopic features among the atypical naevi with different MC1R variants
A significant difference was observed between the number of colours in AN from 0 RHC and 2 RHC groups (1.44 ± 0.22 vs 1.37 ± 0.25, p = 0.007). Adjusting for differences in the number of AN per patient had no influence on the level of significance, although different p-values were observed (Table II). No significant differences in asymmetry, border and structures were observed between 0 RHC variants and 2 RHC variants.
The difference in the total number of colours was due to significantly more dark brown colour in individuals with 0 RHC variants (89% vs 76%, p < 0.001) (Table III). No differences between 0 RHC and 2 RHC variants were observed for the colours light brown, red, blue-grey and black, and no AN with a white colour were observed (Table III). Naevi from individuals with 0 RHC variants contained significantly more streaks (72% vs 52%, p < 0.001) and pigment networks (47% vs 36%, p = 0.023), while naevi from individuals with 2 RHC variants contained significantly more structureless areas (76% vs 95%, p < 0.001) (Table IV). Adjustment in the analyses for inter-patient differences in the number of AN had no influence on the level of significance for dark-brown colour and streaks, while the significant differences for pigment network disappeared (p = 0.556). No significant differences were observed for dots and globules. Almost all 876 naevi contained dots (98%), while globules were only observed in 78 naevi (9%).
No significant difference in mean TDS between 0 RHC variants and one RHC variant was observed. No difference in mean± SD ABCD TDS between naevi containing 0 RHC variants and 2 RHC variants was observed (4.73 ± 1.13 vs. 4.72 ± 1.27, p = 0.96) (Table II).
Table II. Mean ABCD total dermoscopy score (TDS) in 876 clinical atypical naevi from 111 patients according to MC1R red hair colour (RHC) variants
|
ABCD TDS |
0 RHC |
1 RHC |
2 RHC |
p-valuea |
p-valueb |
|
Asymmetry |
1.76 |
1.54 |
1.89 |
0.18 |
0.672 |
|
Borders |
0.007 |
0.003 |
0.007 |
0.96 |
0.969 |
|
Colours |
1.44 |
1.43 |
1.37 |
0.007 |
0.035 |
|
Structures |
1.52 |
1.53 |
1.45 |
0.135 |
0.327 |
|
Total TDS |
4.73 |
4.51 |
4.72 |
0.96 |
0.655 |
aStudent’s t-test.
bLinear Mixed Model, taking in account the inter-patient differences in amount of naevi.
Significant values are shown in bold.
Table III. Percentage of colours among 0 red hair colour (RHC), 1 RHC and 2 RHC variants
|
Colours |
0 RHC n = 513 |
1 RHC n = 253 |
2 RHC n = 110 |
p-valuea |
p-valueb |
|
White |
0 |
0 |
0 |
1.000 |
1.000 |
|
Red |
96 |
96 |
95 |
0.604 |
0.894 |
|
Light brown |
99 |
99 |
100 |
0.613 |
0.613 |
|
Dark brown |
89 |
85 |
76 |
0.000 |
0.009 |
|
Black |
5 |
6 |
3 |
0.449 |
0.196 |
|
Blue |
0 |
0 |
1 |
0.177 |
0.177 |
aSingle χ square test.
bGenLin Model, taking in account the inter-patient differences in amount of naevi.
Table IV. Percentage of structures among 0 red hair colour (RHC), 1 RHC and 2 RHC
|
Structures |
0 RHC n = 513 |
1 RHC n = 253 |
2 RHC n = 110 |
p-valuea |
p-valueb |
|
Dots |
98 |
98 |
97 |
0.722 |
0.604 |
|
Globules |
10 |
6 |
11 |
0.760 |
0.845 |
|
Streaks |
72 |
74 |
52 |
0.000 |
0.000 |
|
Structureless area |
76 |
86 |
95 |
0.000 |
0.000 |
|
Pigment network |
47 |
43 |
36 |
0.023 |
0.556 |
aSingle χ square test.
bGenLin Model, taking in account the inter-patient differences in amount of naevi.
Dermoscopic features among melanomas with different MC1R variants
According to the pathology report, 13 superficial spreading malignant melanomas (SSM) and 8 melanomas in situ (MIS) from CDKN2A mutation carriers were available (Table SI; available from http://www.medicaljournals.se/acta/content/?doi=10.2340/00015555-1457). The melanomas were diagnosed in 17 individuals (12 women and 5 men) heterozygous for the CDKN2A founder mutation and one male (individual 9) homozygous for the CDKN2A founder mutation. Nine persons were carrying 0 RHC variants, 6 persons were carrying 1 RHC variant and 2 persons were carrying 2 RHC variants. The TDS did not differ between 0 RHC variants and 2 RHC variants (6.71 ± 0.92 vs 7.00 ± 0.85). Also, no difference was observed between 0 RHC variants and 1 RHC variant (6.71 ± 0.92 vs 7.18 ± 0.64). Since SSM as well as MIS were included in this study population, the mean TDS for both groups were calculated. No differences in mean TDS was observed between SSM and MIS (7.08 ± 0.70 vs 6.76 ± 0.90).
DISCUSSION
We examined the association between RHC variants and the TDS (and the individual items that make up the TDS score) in AN and melanomas from a group at high risk of melanoma.
Significantly more total colours were observed in AN from individuals with 0 RHC variants, apparently due to the more frequent presence of a dark brown colour in these AN. Still, a dark brown colour was observed in 76% of the AN in individuals with 2 RHC variants, which could be considered high. This relatively high score of dark brown colour in AN from individuals with 2 RHC variants can be explained by the way a colour is scored in the ABCD system. The presence of a minor amount of dark brown pigment already scores, while the surface of the dark brown area is not taken into account.
Streaks and a pigment network were more frequently seen in individuals with 0 RHC variants compared to individuals with 2 RHC variants, while a structureless area was more frequently observed in individuals with 2 RHC variants. Possibly, the presence of a pigment network and a structureless area are inversely correlated, since a pigment network gives a structure to a naevus. Despite the differences in colours between AN from individuals with 0 RHC variants and 2 RHC variants, we did not observe a difference in TDS scores between AN from individuals with 0 RHC variants and 2 RHC variants. However, clinicians should be aware of a different colour and structure pattern between individuals with and without red hair, since a dark-brown colour, streaks and a pigment network are less frequently observed in naevi from persons with 2 RHC variants (Fig. 2).
With our study we were able to test previous findings by Cuéllar et al. (19), who showed that melanomas from CDKN2A mutation carriers with 2 RHC variants had a significantly lower TDS score. We could not confirm these previous findings in 21 melanomas from 17 CDKN2A founder mutation carriers (Fig. 3). In the current study, the mean TDS score in the 2 individuals with 2 RHC variants was even higher than the mean TDS from 9 individuals with 0 RHC variants, although not enough individuals were included to enable statistical analyses. The higher TDS among carriers of 2 RHC variants might be caused by the late detection of the melanoma in patient 3 (SSM, Breslow 1.3, Clark IV). This discrepancy with previously described observations in melanomas from the Spanish CDKN2A mutation carriers might also result from a rather small sample size in both studies, and differences in the selection of high risk patients.
For research study purposes, the use of the TDS/ABCD scoring system is a more useful tool in the evaluation of melanocytic lesions than the more intuitive types of pattern recognition (e.g. Pattern Analyses), since it provides a reproducible score and can be used for numerical comparisons. Although the ABCD TDS is useful in study settings to investigate differences between individuals with different phenotypes, the additional value for persons with lightblond/reddish hair is low. For clinicians, other factors like atypical blood vessels, changes over time and the relation with the patient’s other melanocytic lesions, are even more important than a single TDS.
Fig. 2. Two representative examples of atypical naevi from two CDKN2A mutation carriers were selected as illustration. Naevus 1 (a–c) was selected from a patient with 0 red hair colour (RHC) variants, while naevus 2 (d–f) was selected from a patient with 2 RHC variants. The figure illustrates the prominent dark-brown colour and structures in naevi from individuals with 0 RHC variants compared to the more frequently occurrence of structureless area in naevi from individuals with 2 RHC variants.
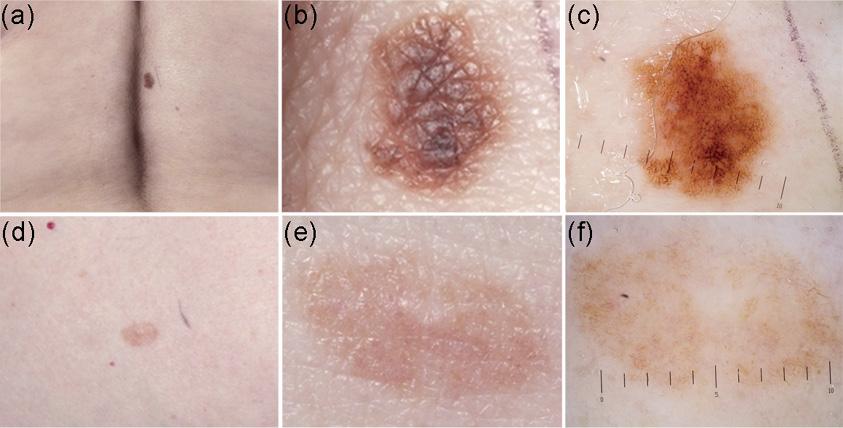
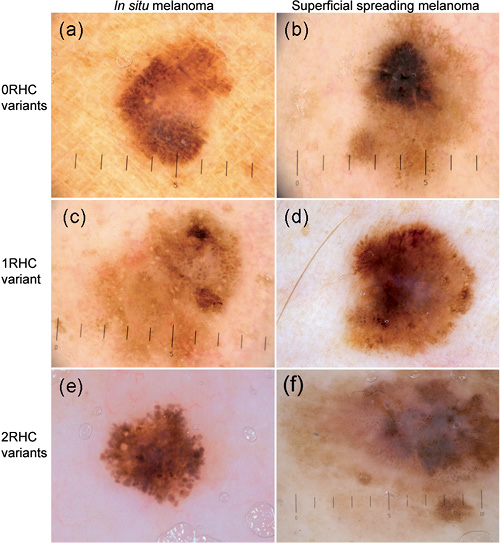
Fig. 3. Examples of three melanoma in situ (a, c, e) and three superficial spreading melanomas (b, d, f) from patients with 0 red hair colour (RHC) variants (a, b), 1 RHC variant (c, d) and 2 RHC variants (e, f). In individuals with 0 RHC variants, a Total Dermoscopy Score (TDS) of 7.4 was observed in the melanoma in situ (a), while the more invasive superficial spreading melanoma (SSM) (b; Breslow 0.66 Clark 3) has a TDS of 7.6. The images of a melanoma in situ (c) and SSM (d; Breslow 0.40, Clark 2) from 2 individuals with 1 RHC variant (c, d) have respectively both a TDS of 7.4. Among the individuals with 2 RHC variants a TDS of 6.4 was observed in the melanoma in situ (e) and 7.6 in the SSM (f; Breslow 1.3, Clark 4). However, also melanomas from 2 RHC variants contain a dark-brown or black colour (×10 magnification).
In the current study the association between RHC variants and ABCD TDS was studied in a group at high risk of melanoma. Although CDKN2A mutation carriers have a higher naevus number and density, no influence on the individual naevus phenotype has been observed (22). We suggest that the current results may be extrapolated to the general population with light blond or red hair.
In conclusion, we demonstrated specific dermoscopic colour and structure differences between AN from patients with 0 and 2 RHC variants. Clinicians should be aware of the different dermoscopic naevus phenotypes in patients with light blond or red hair. Although we observed no difference in TDS between individuals with 0 RHC and 2 RHC variants, we suggest that the TDS is not fully suitable for patients with a very light complexion. The TDS gives an incomplete view, ignoring other important factors (history, naevi phenotype, atypical vessels). Secondly, we did not observe differences in TDS in melanomas from patients with 0 RHC variants and 2 RHC variants since all melanomas, independent of the patient’s RHC status, have a high TDS (almost reaching the upper limit of the TDS). Future studies need to examine our conclusions in those with red hair in the general population.
ACKNOWLEDGEMENTS
We would like to thank Paul Douw van der Krap for taking the clinical images.
This work was supported by the National Cancer Institute (NCI) of the US National Institutes of Health (NIH), contract number CA83115.
The authors declare no conflict of interest.
REFERENCES
