Hans B. LOMHOLT and Mogens KILIAN
Department of Biomedicine, Aarhus University, Aarhus, Denmark
Increasing antibiotic resistance in the population of Propionibacterium acnes is a major concern. Our aims were to examine the clonal relationships and anatomical distribution of resistant and sensitive P. acnes. A collection of 350 P. acnes isolates was therefore used to determine the minimum inhibitory concentration of tetracycline, erythromycin and clindamycin, multilocus sequence type, and the identity of genetic resistance markers. Two hitherto unknown resistance mutations were detected. Resistant P. acnes mainly belonged to clonal clusters in division I-1a frequently isolated from skin and associated with moderate to severe acne. All high-level tetracycline resistant strains were members of a single clone. Multiple isolates from distinct anatomic areas of surface skin and follicles of 2 acne patients revealed substantial clonal diversity between areas and co-existence of resistant and sensitive clones. Fifty-two percent of Danish acne patients and 43% of controls carried at least one resistant P. acnes strain, resistance to clindamycin being most frequent followed by tetracycline and erythromycin. Resistance to tetracycline was detected exclusively among isolates from acne patients. In conclusion, antibiotic resistance is associated with particular evolutionary clades of P. acnes and a substantial part is due to a single geographically widespread clone (ST3). Individuals carry a strikingly complex population of P. acnes with distinct virulence potential and antibiotic resistance. Key words: Propionibacterium acnes; antibiotic resistance; acne; multilocus sequence typing; clonal diversity.
Accepted Nov 7, 2013, Epub ahead of print Feb 27, 2014
Acta Derm Venereol
Hans B. Lomholt, Department of Biomedicine, Aarhus University, Wilhelm Meyers allé 4, DK-8000 Aarhus C, Denmark. E-mail: hans.lomholt@dadlnet.dk
Antibiotics have been the mainstay of acne treatment for many years. The necessity for long treatment periods makes bacterial resistance a major concern. Propionibacterium acnes resistance to erythromycin was described as early as 1972 (1). Since then widespread resistance among P. acnes to macrolides, lincosamines, and tetracyclines has been reported from many countries worldwide (1–10). Resistance in P. acnes to relevant antibiotics is due mainly to a few point mutations in genes encoding ribosomal RNAs. While resistance to tetracyclines is due to a single mutation in the 16S rRNA gene (1058G>C), resistance to MLS antibiotics (macrolide-lincosamide-streptogramin B, including erythromycin and clindamycin) is associated with 4 genotypic groups I to IV defined by 3 different mutations in the 23S rRNA gene (group I: 2058A>G; group III: 2057G>A; group IV: A2059A>G) or presence of the RNA methylase gene erm(X) (group II) on a mobile genetic element (8, 11).
The clinical impact of P. acnes resistance on the acne treatment outcome has been surprisingly difficult to estimate although a few studies have indicated that patients harbouring resistant P. acnes may respond less well to antibiotic treatment (2, 12–14). A review of clinical trials of topical formulations of erythromycin and clindamycin showed a significant gradual decrease in the efficiency of topical erythromycin against inflammatory and non-inflammatory acne lesions since its introduction in the mid-1970s (15).
The genetic heterogeneity of P. acnes has become increasingly clear (16, 17). Recently, the population structure of P. acnes was disclosed in detail by use of multilocus sequence analysis (MLSA). Interestingly, certain genetic lineages of P. acnes were found to be significantly associated with moderate to severe acne while others were associated with healthy skin (16, 17). Only sparse information is available on the distribution of resistance genes within the P. acnes population. Oprica et al. (18) showed that tetracycline resistant P. acnes have closely related pulsed field gel electrophoresis (PFGE) patterns while no correlation was detected between 23S rRNA mutations causing clindamycin and erythromycin resistance and PFGE types. Recently, McDowell et al. (19) examined 38 resistant P. acnes strains with rRNA mutations by use of MLSA and found that one clonal lineage dominated among resistant strains. In another recent study, mapping of the skin population of P. acnes based on comprehensive analysis of 16S rRNA sequences revealed 3 P. acnes ribotypes with recognised resistance-associated mutations (20).
The present study was undertaken to examine the relation between resistance genes and evolutionary lineages of P. acnes and the diversity and anatomic distribution of resistant and susceptible clones carried by individuals with acne.
MATERIALS AND METHODS
Propionibacterium acnes strains
A total of 350 P. acnes isolates were included in the study: (i) 180 strains consisting of 114 previously described non-redundant and unselected strains isolated during 2006 and 2007 from 39 Danish subjects, (8 with moderate to severe acne, 17 with mild acne, and 14 with healthy skin) (16), 16 high-level resistant isolates from acne patients in a Danish dermatology practise collected August 2010 to February 2012, 20 strains from healthy skin of Chinese individuals sampled in 2008, and 30 CCUG strains isolated from acne and various opportunistic infections in the United Kingdom, U.S.A., Sweden, Norway, and Germany collected between 1920 to 2004, (ii) 170 isolates of P. acnes recovered from 2 acne patients. Samples collected as described (16) were inoculated on anaerobically incubated TYG agar (20 g/l tryptone, 10 g/l yeast extract, 5 g/l glucose with 1% agar) supplemented with 2 mg/ml furazolidone. From patient A 30 isolates were collected from each of cultures from the face and back, and 20 additional isolates were collected from the face 6 months later. From patient B 90 colonies were isolated from surface skin of the face (n = 30), the upper part of the back (n = 30), and from sebaceous follicles in the face (n = 30).
The study protocol was approved by the Ethics Committee of the County of Aarhus, and the study was conducted according to the principles of the declaration of Helsinki.
Antibiotic susceptibility testing
The minimum inhibitory concentration (MIC) of tetracycline, clindamycin and erythromycin was determined in duplicate for all isolates by an agar dilution method using TYG agar plates containing 2-fold dilutions of the respective antibiotics in concentrations from 0.002 µg/ml to 256 µg/ml. Plates without antibiotics were included to control for growth. The inoculum was approximately 105 colony forming units transferred to the agar surface with a 96 pin applicator from a microtitration plate containing suspensions of the individual strains. The lowest dilution with absence of growth was defined as the MIC. Resistance to tetracycline was defined as MIC ≥ 2 mg/l, to clindamycin MIC ≥0.25 mg/l, and to erythromycin MIC ≥ 0.5 mg/l according to EUCAST (21). High-level resistance was defined for tetracycline as MIC ≥32 mg/l, for clindamycin as MIC ≥128 mg/l, and for erythromycin as MIC ≥256 mg/l.
Sequence analysis of 16S and 23S rRNA genes and PCR detection of erm(X)
PCR amplification of relevant 16S and 23S rRNA and erm(X) gene sequences was performed as described by Oprica et al. (18) Presence of erm(X) was visualised as an amplicon of appropriate size in 2% agarose gels. Arcanobacterium pyogenes 98-4277-2 donated by Carl-Erik Nord, Karolinska Institutet, Stockholm, Sweden, served as positive control.
Purification of PCR amplicons was performed on Wizard Minicolumns (Promega, Madison, Wis.). Sequencing of the amplified fragments in both directions was achieved with the PCR primers and the Thermo Sequenase dye terminator cycle sequencing kit (Amersham Life Science) on Applied Biosystems automated sequencers 3730xl.
Multilocus sequence typing
MLST using the Aarhus scheme (16, 22) was based on 9 housekeeping genes with reference to the database at http://pacnes.mlst.net/. Clonal clusters (CC) were identified with the eBURST v.3 software at http://eburst.mlst.net according to the principles described by Feil et al. (23). Sequences were inspected in MEGA version 5.05 (24).
Statistical analysis
Differences between proportions of resistant isolates from healthy subjects and acne patients and between proportions of carriers of resistant strains were statistically evaluated by Fisher’s exact test (2-tailed) performed in InStat.
RESULTS
Antibiotic resistance among P. acnes isolates
MIC values for the 114 unselected Danish P. acnes isolates (72 from acne patients and 42 from healthy controls) are shown in Fig. S11. According to the defined breakpoints, 15.8% were resistant to clindamycin (3.5% high-level), 8.8% were resistant to erythromycin (5.5% high-level), and 9.6% were resistant to tetracycline (1.8% high-level). Among the 7 high-level resistant isolates, 6 were from acne patients, and the 7th was from a healthy dermatologist attending acne patients.
As shown in Fig. S11, the distribution of MIC values of clindamycin and erythromycin did not differ between acne and control isolates. In contrast, a significantly higher proportion of isolates from acne patients than control isolates were resistant to tetracycline (p = 0.0066).
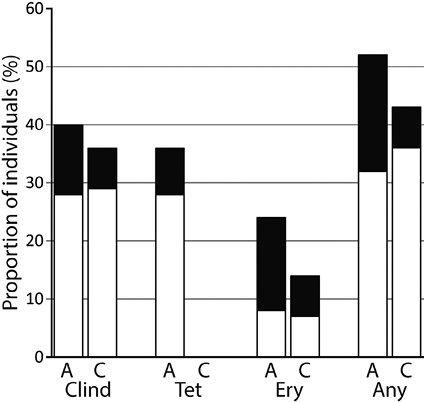
The proportion of the 39 Danish individuals that carried resistant isolates is shown in Fig. 1. Among the 25 acne patients 52% carried at least one strain resistant to one of the 3 antibiotics compared to 43% of the 14 controls (p = 0.4393). Tetracycline resistance was exclusively detected among acne patients.
The proportions of strains with different combinations of antibiotic susceptibility detected in the collection of 114 Danish strains are shown in Table I.
Table I. Combinations of resistance to tetracycline, clindamycin and erythromycin detected among 114 non-redundant Danish P. acnes isolates
|
Antibiotic |
Isolates n (%) |
|
Clindamycin only |
12 (10.5) |
|
Tetracycline only |
6 (5.3) |
|
Erythromycin only |
3 (2.6) |
|
Clindamycin-Tetracycline |
3 (2.6) |
|
Clindamycin-Erythromycin |
3 (2.6) |
|
Tetracycline-Erythromycin |
0 |
|
Clindamycin-Erythromycin-Tetracycline |
5 (4.4) |
Among 20 isolates from healthy carriers in Beijing, China, 3 showed low-level resistance to clindamycin. Among the 30 CCUG strains one strain was resistant to erythromycin and 2 other strains were resistant to clindamycin. None of these strains showed high-level resistance and none was resistant to tetracycline.
Resistance in relation to molecular mechanism and population structure
The relationships between sequence type defined by allelic profile of 9 housekeeping genes, genetic clades, and the degree and mechanism of high-level resistance are shown in Table SI1.
High-level resistance was mainly associated with isolates of clone ST3 of clonal complex 3 (CC3) and three STs representing CC18. The ST3 strains were isolated from acne patients attending 3 independent dermatology clinics in Denmark. Notably, high-level tetracycline resistance was exclusively detected in this clone.
Interestingly, one CC36 and one ST52 (type II) strain showed 2 different and not previously recorded mutations in the 23S rRNA gene at position 2058 (A>T and A>C), both associated with high-level erythromycin resistance and low-level clindamycin resistance.
The genetic relations of low-level resistant isolates are shown in Table SII1. Like the high-level resistant isolates, most low-level resistance was detected in isolates assigned to CC3 and CC18. No erm(X) or mutations in 23S rRNA and 16S rRNA genes were detected in low-level resistant isolates.
Clonal diversity and anatomic distribution of P. acnes strains with distinct susceptibility patterns
The analyses of 114 isolates from 39 Danish subjects showed that single individuals carried 1–6 clones of P. acnes with distinct patterns of susceptibility to antibiotics. To add further detail, we performed a comprehensive analysis of the clonal distribution and antibiotic susceptibility of the P. acnes population on the skin of 2 acne patients. Based on different patterns of antibiotic susceptibility representative isolates were selected for MLST and analysis of genetic determinants of resistance and results were translated to the complete set of isolates as shown in Fig. S21.
Patient A was sampled from surface skin of the face (Leeds score 6) and back (Leeds score 0) and again from the face 6 months later (Leeds score 0). Treatment with isotretinoin was initiated after collection of the initial samples. While a susceptible ST18 clone dominated facial skin, an ST20 clone with high-level erythromycin resistance dominated the skin of the back. Notably, the clonal distribution of the P. acnes population on the face appeared to be almost unaltered after 6 months of isotretinoin treatment.
Patient B had moderate acne (Leeds 4 in the face and < 1 on the back) 6 months before the bacteriological sampling and was treated with tetracycline for 6 months. At the subsequent bacteriological sampling no active acne was observed. Four different clones were identified including 2 with high-level erythromycin resistance. All isolates of the ST3 and the ST29 clone were analysed for mutations in the 16S rRNA gene to distinguish these clones with similar resistance patterns. The distribution of clones differed significantly between the 3 anatomic regions. Notably, an antibiotic-susceptible ST31 clone constituted a substantial part of the follicular P. acnes population but was absent on the facial skin surface. Interestingly, the patient carried 2 clonally distinct isolates with the same 2058A>G mutation.
DISCUSSION
The aims of this study were to determine the prevalence, molecular mechanisms, and clonal relationships of antibiotics resistance in P. acnes carried by acne patients and healthy controls. The study focused on 3 antibiotics: tetracycline, clindamycin and erythromycin, which are widely used for treatment of acne.
Our MLST analysis of resistant isolates provides interesting information on the molecular epidemiology of resistance in the P. acnes population. Thus, the majority of both high- and low-level resistant P. acnes isolates belonged to 2 clonal clusters of phylogenetic division I-1a (Tables SI1 and SII1). This is consistent with a recent report by McDowell et al. (19) and with the lack of resistance among isolates from prostate tissue and gastric mucosa, which are dominated by type II strains (25, 26).
Although our isolates were from a relatively geographically restricted area, the results provide important information on the epidemiology of resistance when combined with recent reports. A single clone, ST3, was markedly overrepresented among resistant strains (Table SI1). A recent comprehensive 16S rRNA-sequence based study from the United States identified the same genotype (designated RT4) strongly associated with acne, though present only in a fraction of the examined patients (20). Likewise, McDowell et al. (19) recently reported that the same ST3 clone with one or both 16S rRNA mutations constituted 52% of 38 resistant strains, including isolates from Italy, Sweden, the U.K., Australia, and the U.S.A. Taken together, these findings demonstrate that this resistant ST3 clone successfully spread in the Western world and accounts for a major part of high-level resistant P. acnes. The observation by McDowell et al. that 24% (5/21) of their resistant CC3 isolates were from healthy skin of individuals not treated with antibiotics show that the clone may be common also in some healthy populations dependent on local antibiotic usage.
Our previous observations, subsequently supported by others, show that certain evolutionary lineages of P. acnes, particularly ST18, are associated with acne while others are associated with health and even may be beneficial (16, 17, 20). In this context, it is intriguing that our detailed sampling of 2 patients revealed substantial anatomical differences in the clonal distribution and antibiotic susceptibility of P. acnes carried by single individuals (Fig. S21). Thus, clones prevalent in the follicles may not be prevalent on the skin surface. This complexity complicates evaluation of the relevance of resistance for the clinical treatment outcome.
High-level resistance to erythromycin, often associated with high-level clindamycin resistance, was detected in several clonal clusters including the acne-associated CC18. This would explain why P. acnes resistance is associated with a reduced effect of erythromycin treatment (12, 15).
The majority of resistance was associated with previously demonstrated rRNA genotypes and in relative frequencies similar to previous reports (18, 19, 27, 28). However, 2 isolates showed 23S rRNA gene mutations not previously described but both associated with high-level erythromycin resistance and low-level clindamycin resistance. In addition, one isolate highly resistant to erythromycin had neither mutations in the 23S rRNA gene nor evidence of erm(X), which corresponds to recent observations for 2 Swedish isolates (19). These data illustrate the emergence of alternate and yet unidentified mechanisms of high-level resistance to MLS antibiotics.
The detected prevalence of resistance among Danish isolates is in the lower end of the spectrum recorded for other European countries (4) consistent with a relatively restricted use of antibiotics in Denmark. The striking finding that high-level tetracycline resistant P. acnes were carried exclusively by acne patients concurs with the almost exclusive use of tetracycline for the treatment of acne in Danish primary health care (29). In a comparable Swedish study, Oprica and co-workers (5) detected tetracycline resistant P. acnes in only 14% of acne patients. Yet, the consumption of tetracycline in Sweden is significantly higher than in Denmark (3.44 defined daily doses versus 1.48 defined daily doses per 1,000 and day in 2007) but a significant part is used in the age group 60–79 years for treatment of patients with chronic respiratory infections and not related to acne (29, 30). These findings support the conclusion that development of high-level tetracycline resistance in P. acnes and sustained carriage is primarily associated with prolonged acne treatment regimens.
In conclusion, resistance to antibiotics relevant to treatment of acne is associated with particular evolutionary lineages of P. acnes including a particular clone (ST3) that shows geographically widespread dissemination. Individuals may carry a strikingly complex population of P. acnes with diverse virulence potential and antibiotic resistance patterns. These findings provide additional explanation for the difficulties in predicting the clinical effects of antibiotic treatment of acne. Theoretically, selection of a resistant non-virulent clone may result in clinical improvement.
ACKNOWLEDGEMENTS
This study was supported by Fonden for Faglig Udvikling af Speciallægepraksis.

1http://www.medicaljournals.se/acta/content/?doi=10.2340/00015555-1794
REFERENCES
