Xiaodong ZHANG1,2#, Weijun LIU1,2#, Weida LIU1,2, Haiqin JIANG1,2, Wenkai ZONG1, Guoyi ZHANG1, Peiying JIN1 and Hongsheng WANG1,2
1Institute of Dermatology, Chinese Academy of Medical Sciences and Peking Union Medical College, and 2Jiangsu Key Laboratory of Molecular Biology for Skin Diseases and STIs, Nanjing, China
#These authors contributed equally to this work and should be considered as first authors.
Skin and soft tissue infections caused by rapidly growing non-tuberculous mycobacteria (RG-NTM) have become a growing clinical concern over the past decades. These RG-NTM are ubiquitous environmental organisms and most are resistant to traditional antituberculous agents. In this report, we describe 3 cutaneous infections caused by RG-NTM, namely, Mycobacterium abscessus, M. chelonae, and M. conceptionense, and present the clinical and laboratory characteristics of these infections. Key word: rapid mycobacterial growth; Mycobacterium abscessus; Mycobacterium chelonae; Mycobacterium fortuitum; Mycobacterium conceptionense.
Accepted Mar 19, 2015; Epub ahead of print Mar 25, 2015
Acta Derm Venereol
Hongsheng Wang, Institute of Dermatology, Chinese Academy of Medical Sciences and Peking Union Medical College, Jiangsu Key Laboratory of Molecular Biology for Skin Diseases and STIs, 12 Jiangwangmiao Road, Nanjing 210042, China. E-mail: whs33@vip.sina.com;
Non-tuberculous mycobacteria (NTM) are responsible for a broad spectrum of human diseases, that may present with localized, regional or disseminated infections (1). NTM have been grouped into 4 broad categories according to the Runyon system, based on growth rates and pigment patterns (2). Rapidly growing NTM (RG-NTM) comprise Runyon group IV, which are mycobacteria that form colonies in subcultures within 7 days, in contrast to slower-growing mycobacteria (Runyon groups I–III) that require long incubation periods (3). RG-NTM are increasingly recognized as important human pathogens that cause various diseases ranging from localized cutaneous infections to disseminated infections; haematogenously disseminated infections are usually seen only in immunocompromised hosts (2, 4, 5). The majority of infections are due to accidental inoculation from trauma, surgery, injection, or cannulation. Pulmonary infection is usually associated with underlying structural lung disease. Although nearly a hundred of RG-NTM have been identified, Mycobacterium abscessus, M. chelonae, and M. fortuitum are the most common species that are pathogenic. In this report, we describe 3 immunocompetent hosts with cutaneous infections caused by M. abscessus, M. chelonae, and M. conceptionense; we review the literature of management of such cutaneous infections.
CASE REPORTS and Methods
Case 1. A previously healthy 40-year-old woman presented with a 9-month history of deep erythematous, subcutaneous nodules, abscesses, and skin ulcers on the flexor side of her lower legs. These lesions initially appeared as erythema and papules on the left calf, and they gradually extended to the right lower leg. The patient was diagnosed with nodular vasculitis at a local hospital and treated with a combination of glucosidorum tripterygll totorum (Chinese traditional herb), dipyridamole tablets, dirithromycin, and surgical dressings. After 6 months of treatment, no significant therapeutic effects were found. The patient was then transferred to our hospital. Since the onset of the disease, the patient did not complain of any systemic discomfort. Dermatological examination revealed a large area of deep erythema and edema with nodules, ulceration, and abscesses on the right and almost the whole left calves (Fig. 1A). A biopsy taken before admission displayed inflammatory granulomas, without caseous necrosis.
Case 2. A 50-year-old woman was referred to our hospital with a 4-month history of two asymptomatic purple subcutaneous nodules covered with scabs and scales on the extensor surface of her lower leg. These lesions evolved from a millet grain-sized red papule. The patient did not report any trauma. She did not complain of any discomfort. Dermatological examination showed two purple bean-sized nodules covered with thin scales and crust on the extensor of her left shin (Fig. 1C). A skin biopsy of the lesion revealed granulomatous inflammation without caseous necrosis (Figs. 1E and 1F).
Case 3. A 67-year-old woman presented with asymptomatic swollen reddish plaques on her right foot for two months. The lesions first appeared on her right foot after being bruised by wood, and gradually developed furher. Since the onset of the disease, the lesion had never disappeared and the patient did not complain of any systemic discomfort. Dermatological examination showed a thumb-sized reddish nodule, which was smooth and soft without ulcer or scale, on the dorsum of her right foot (Fig. 1G). Biopsy specimens obtained from the lesions were stained with hematoxylin and eosin. An epidermoid cyst containing necrotic material and red blood cells, surrounded by epithelioid cell granulomas, was found (Fig. 1I and J).
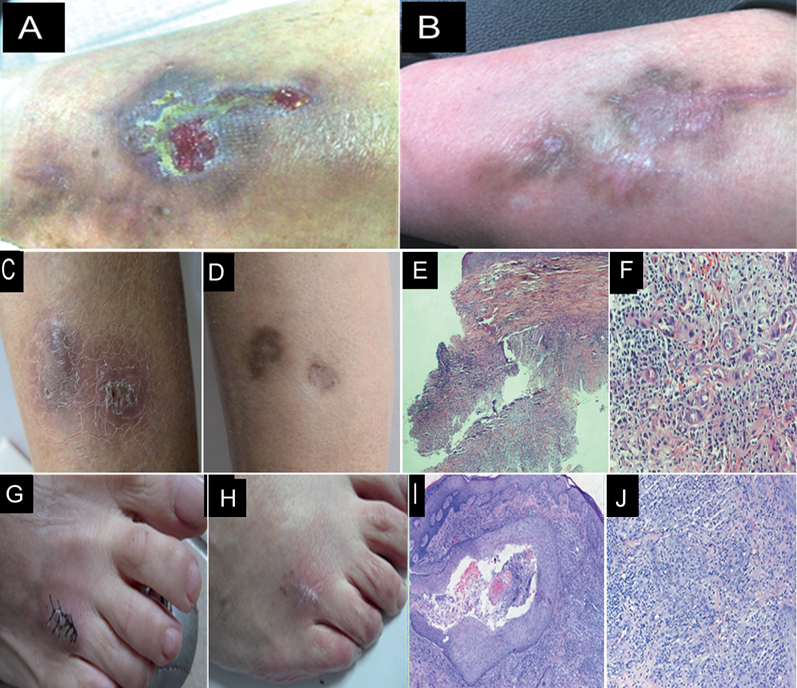
Fig. 1. Erythemas, skin ulcers on the flexor side of left low leg of Case 1 (A). After 6 months of therapy the lesions on patient 1 subsided (B). Nodules on the extensor surface of the lower leg in case 2 (C). After 6 months of therapy the lesions of Case 2 improved (D) (E, F) Histopathological findings of case 2: inflammatory granuloma without caseous necrosis, original magnification ×40,×100; (G) lesions on the right foot of Case 3 after skin biopsy. (H) After 3 months of therapy the lesions of Case 3 were healing; (I, J) Histopathological findings of case 2: Epidermoid cyst surrounded by epithelioid cell granulomas, original magnification ×40, ×100.
All the patients were in good general condition. Routine laboratory analysis presented no abnormalities. No active pulmonary disease was detected by chest radiography. No patients had received any immunosuppressant therapy.
Specimen preparation, microscopy and culture procedure. Skin tissue homogenates were inoculated on Löwenstein–Jensen (L–J) medium and separately incubated at 4 different temperatures (28°C, 32°C, 37°C, and 45°C). Ziehl–Neelsen staining was used to detect smears from skin tissue homogenates and confirm whether the cultured productions are acid-fast bacilli (AFB). Pigmentation testing was conducted for the isolated strains. Fungal and other standard bacterial cultures were also prepared. In vitro drug susceptibility testing was performed on the strain isolated in accordance with Clinical and Laboratory Standards Institute guidelines (6–10).
Preparation of template DNA. One loop of bacteria was harvested from each L–J medium slope incubated at 37°C and then suspended in 180 μl of sterile distilled water. Samples were frozen in liquid nitrogen and transferred to boiling water 5 times to release mycobacterial DNA. After centrifugation to pellet insoluble debris, the supernatants were used as DNA templates for PCR (6–10).
PCR, PCR-restriction fragment length polymorphism (RFLP) and gene sequencing. Two oligonucleotide primers (forward, 5’-ACCAACGATGGTGTGTCCAT-3’; reverse, 5’-CTTGTCGAACCGCATACCCT-3’) were used to amplify a 439 bp fragment of the mycobacterial hsp65 gene. Two other oligonucleotide primers (forward, 5’-GAGATACTCGAGTGGCGAAC-3’; reverse, 5’-GGCCGGCTACCCGTGGTC-3’) were used to amplify a 208 bp fragment of the mycobacterial 16s rRNA sequence. PCR was performed in a 50 μl reaction volume containing 10 μl of the DNA template, 1.25 U of Taq polymerase (Promega Corporation, Madison, WI, USA), 10× reaction buffer, 200 μmol/l deoxyribonucleoside triphosphate, 2.5 mmol/l magnesium chloride, and 1 μmol/l each primer. The thermal profile for amplification of the mycobacterial hsp65 gene involved initial denaturation at 94°C for 5 min; 45 cycles of 94°C for 1 min, 60 °C for 1 min, and 72°C for 1 min; and finally, an extension of 10 min at 72°C. The PCR conditions for amplification of the mycobacterial 16s rRNA sequence involved incubation for 10 min at 40°C and 40 cycles of 94°C for 1.5 min, 65°C for 2 min, and 72°C for 3 min. After the last PCR cycle, the vials were maintained at 72°C for 10 min. The presence of amplified products was confirmed by agarose gel electrophoresis (6–11).
The strains were initially identified by PCR-RFLP analysis, and the results were analyzed by the PCR-RFLP database (http://app.chuv.ch/prasite/index.html). The PCR products of the hsp65 and 16s rRNA genes were also sequenced, and Basic Local Alignment Search Tool (BLAST) program was used to compare the sequences with those in the GenBank database (http://www.ncbi.nlm.nih.gov/BLAST) (11–13).
RESULTS
The results of Ziehl–Neelsen staining of tissue homogenates from 3 cases were negative. Rapid growth of the smooth colonies without pigmentations were noted within 7 days (mean 5 days) of incubation at 28°C, 32°C, and 37°C. No colonies grew at 45°C. Ziehl–Neelsen staining demonstrated that all the cultured organisms were AFB. Data of drug susceptibility testing in vitro of the 3 isolates are shown in Table SI1.
A fragment encoding mycobacterial 65 kDa hsp (hsp65) was amplified in Isolate 1 from Case 1. Digestion of the PCR product yielded two fragments of 235 and 210 bp with BstEII, and 4 fragments of 145, 70, 60, and 55 bp with HaeIII. The RFLP pattern of the isolated strain was identical to that of M. abscessus. Sequencing of the hsp65 gene and 16s rRNA showed 100% homology with M. abscessus. For Isolate 2 from Case 2, the PCR product of hsp65 from the isolate was digested. Digestion yielded two fragments of 320 and 130 bp with BstEII, and 4 fragments of 200, 60, 55, and 50 bp with HaeIII. The RFLP pattern of the isolated strain was identical to that of M. chelonae. Sequencing of the hsp65 gene and 16s rRNA showed 100% and 99% similarity to the M. chelonae, respectively. Sequencing of the hsp65 gene and 16s rRNA of Isolate 3 from Case 3 demonstrated 99% and 100% homology with M. conceptionense, respectively, which was supported by the results of PCR-RFLP pattern analysis: A fragment of 65 kDa hsp, was amplified in Isolate 3. Digestion of the PCR products yielded two fragments of 235, 120 and 85 bp with BstEII, and 4 fragments of 140, 125, 60, and 55 bp with HaeIII.
Case 1 was treated with orally administered clarithromycin (500 mg, twice daily) and moxifloxacin (400 mg, once daily) for one month. The lesions noticeably improved. Based on the results of drug susceptibility testing in vitro (Table SI1), the therapeutic regimen was then altered with the addition of rifabutin (150 mg, once daily). However, the leukocyte levels decreased to 2 × 109/l, which was considered to be an adverse reaction of rifabutin. Administration of all the above-mentioned drugs was stopped, and 2-(2-Ethoxy-2-oxo-1- phenylethyl)-1, 3-thiazolidine-4-carboxylic acid (Leucoson) (20 mg, 3 times daily) were given to the patient. The number of leukocytes increased after 3 days and then normalized after one week. The patient was then treated with orally administered clarithromycin and moxifloxacin again. The patient recovered after almost 6 months of treatment (Fig. 1B). Case 2 was empirically treated with clarithromycin and moxifloxacin in the same doses as Case 1 for one month. The results of drug susceptibility testing show that the two drugs were sensitive to the strain, so the regimen continued. After 6 months of treatment the patient improved. The patient responded well to the therapy, with gradual resolution of the lesions in 6 months. Based on the results of drug susceptibility testing, Case 3 was treated for 3 months with oral clarithromycin and moxifloxacin. The lesions generally disappeared, but hyperpigmentation and scars were left (Fig. 1G).
DISCUSSION
M. abscessus, M. chelonae, and M. fortuitum are the most common pathogenic RG-NTM species that can cause invasive skin and soft tissue infections, as well as pneumonia. Similar to other NTMs, they are opportunistic pathogens that are common in the environment. Given that they are relatively resistant to many common disinfectants, such as chlorine, organomercurials, and glutaraldehyde, they can survive in treated water sources (13, 14). Infections can be caused by non-sterile techniques or contaminated materials following surgery, liposuction, foreign body implantation, mesotherapy, acupuncture, and soft tissue augmentation (15). Although systemic and pulmonary infections are more likely to be contracted by people with immunodeficiency, localized cutaneous infections occur in the healthy. The clinical symptoms of RG-NTM infections vary, ranging from asymptomatic to tender erythematous violaceous nodules, cellulitis, ulcer, and draining sinuses with discharge. Our cases who were immunocompetent, presented with plaques, nodules and ulceration limited to skin. Only one of them had a history of trauma.
M. chelonae was first identified in 1903 when Friedman isolated an AFB from the sea turtle Chelonae corsicata, and it has long been confused with M. abscessus (16). Other closely related species include M. massiliense, M. bolletii, M. salmoniphilum, M. immunogenum, and M. franklinii. All these species form the M. chelonae-abscessus complex, which is the 3rd most common pathogenic mycobacterial complex apart from the M. tuberculosis and M. avium complexes because of their similar phenotypes. DNA sequencing (e.g., hsp65, rpoB and secA) or PCR restriction enzyme analysis on a population of isolates enabled us to identify them (17–20). M. abscessus is part of an RG-NTM group that includes at least 3 subtypes, namely, M. abscessus, M. massiliense, and M. bolletii. This group was proposed based on phylogenetic trees constructed with data from whole genome sequencing and single nucleotide polymorphism information from the core genomes. The subtypes of M. abscessus share 99% sequence identity with one another, and they cannot be clearly discriminated by phenotypic or molecular tests (21). Over the past few decades, outbreaks and isolated cases of skin and soft tissue infections caused by M. chelonae and M. abscessus have been described in association with cosmetic surgery and tattooing (17–22).
M. conceptionense is a new species belonging to the M. fortuitum group. This organism was first isolated from a patient with posttraumatic osteitis in France in 2006. It is an acid-fast and Gram-positive bacillus. Colonies are non-pigmented and appear on egg-based L–J slants in 2–5 days at temperatures between 25°C and 37°C (optimal temperature: 30°C). No growth occurs at 42°C. Water is a proven source for M. conceptionense (23). Clinical reports about this novel organism are lacking. At the time of writing, only 11 cases of M. conceptionense infection have been reported since 2006. Most patients have underlying disease or trauma history, except one. Among these 11 cases, 4 involved skin and soft tissue infections (Table I) (23–29).
Table I. Clinical characteristics of previously reported patients with cutaneous infections due to Mycobacterium conceptionense
|
Patient |
Age, years/Sex |
Trauma or not |
Underlying disease |
Identification methods |
Treatment |
Outcome |
|
|
Drugs |
Duration, months |
||||||
|
Adekambi, et al. (23) |
31/F |
Open right tibia fracture |
No |
16s rRNA, soda, hsp65, recA, rpoB |
Antimicrobial drug therapy; AMC |
3 |
– |
|
Liao, et al. (24) |
43/F |
No |
No |
16SrDNA |
Surgery, antimicrobial drug, COT, CLA, DOX, LIN |
5 |
Success |
|
Thibeaut, et al. (25) |
58/F |
Breast implant infection |
No |
rpo B |
Surgery, antimicrobial drug, CIP and AZY; then CIP, AZY, and DOX; then DOX |
18 |
Success |
|
Yang, et al. (26) |
50/F |
Lipoinjection |
No |
16SrRNA, rpoB |
AMK, LVX, CFX, CLA, SXT |
1 |
Success |
AMC: amoxicillin/clavulanic acid; COT: cotrimoxazole; CLA: clarithromycin; DOX: doxycycline; LIN: linezolid; CIP: ciprofloxacin; AZY: azithromycin; Success: no relapse after at least 2 months follow-up.
Smear preparation is a fast and feasible diagnostic method, but it is not sensitive and cannot identify the species. Culture in vitro is particularly valuable in defining RG-NTM. It is the foundation of drug susceptibility tests and identification. However, it is influenced by contamination originating from the environment. Sequencing of hsp65, which is used in this report, can provide reliable and rapid identification of RG-NTM. Another widely used molecular method for classification and identification of bacteria is 16S rRNA gene sequencing, which cannot identify the subtypes of RG-NTM. Recently introduced molecular methods such as sequencing of rpoB, secA1, internal transcribed spacer of the 16S-23S rRNA gene, superoxide dismutase gene, dnaJ, secA, recA1, dnaK, and the 32 kDa protein gene, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, pyrosequencing, DNA chip technology, and Beacon probe-combined PCR probes are available in the detection of various species of RG-NTM (12, 15–21, 30, 31). However these techniques are not widely utilized in routine practice.
Diseases caused by RG-NTM are intractable, and the recurrence rate of such diseases is high (19). RG-NTM are famous drug-resistant mycobacterial species. M. abscessus might be susceptible to amikacin, clarithromycin, tigecycline, and cefoxitin. Some subspecies of M. abscessus demonstrate at least intermediate susceptibility to linezolid and imipenem. M. chelonae shows excellent in vitro susceptibility to tobramycin, clarithromycin, linezolid, amikacin, and tigecycline, but it is uniformly resistant to cefoxitin (16–22). Treatment for M. chelonae is generally more successful than that for M. abscessus. Members of the M. fortuitum group are typically more susceptible to sulfonamides, ciprofloxacin, imipenem, linezolid, tigecycline, and doxycycline. However, the M. fortuitum group demonstrates variable expression of a gene that induces resistance to macrolides; therefore, the results of susceptibility to macrolides should be interpreted with caution (23–29). However the therapies were different in previous reports. The efficacy is related to the range of infections, systemic immune state of the patients. The choice of antibiotic drugs base on in vitro susceptibility tests, although the results are not equal to efficacy in vivo. That’s why Individual difference should be considered when we decide the therapy. Generally, an antibiotic combination is better than single-drug therapy. Treatment must be administered for a sufficiently long time period to ensure complete lesional healing and no recurrence. In this combined clinical experience with tigecycline-containing regimens for salvage treatment of RG-NTM infections, clinical improvement was evident in 48.1% of patients (4, 5, 10, 15). However, all the cases in this report improved by oral clarithromycin and moxifloxacin.
Conclusion
Diagnosing RG-NTM infections is difficult for lack of characteristic clinical manifestations. Pathological findings of granulomatous inflammation may provide clues of chronic infection but not pathogens. When standard bacterial cultures as well as microscopic examination are all negative, and histopathological examination showed infectious granulomas without caseous necrosis, RG-NTM infection should be considered. Isolating and identifying the organism is significant to diagnosis. Our diagnoses were supported by clinical features, pathological characteristics and species identifications. Because of the relative limitations of current treatment options, we choose drugs based on antimicrobial susceptibility tests. The effectiveness of the treatments confirmed our diagnoses. Our findings may improve knowledge about these diseases and should remind physicians to suspect NTM infections in otherwise healthy patients with chronic cutaneous infections.
ACKNOWLEGEMENTS
This study was supported by grants from the National Natural Science Foundation of China, 2013 (81371751) and Funded by Jiangsu Provincial Special Program of Medical Science (BL2012003). We thank all the medical workers in the hospital for their cooperation and clinical assistance.
The authors declare no conflict of interest.

1http://www.medicaljournals.se/acta/content/?doi=10.2340/00015555-2105
REFERENCES
