Jouni Uitto
Department of Dermatology and Cutaneous Biology, Jefferson Medical College, and Jefferson Institute of Molecular Medicine, Thomas Jefferson University, Philadelphia, Pennsylvania, USA
Jouni Uitto
Department of Dermatology and Cutaneous Biology, Jefferson Medical College, and Jefferson Institute of Molecular Medicine, Thomas Jefferson University, Philadelphia, Pennsylvania, USA
Epidermolysis bullosa, a group of blistering disorders, serves as the paradigm of the tremendous progress made in understanding the molecular genetics of heritable skin diseases. Mutations in 10 distinct genes have been disclosed in the classic forms of epidermolysis bullosa, and the level of expression of the mutated genes within the cutaneous basement membrane zone, the types and combinations of mutations and their consequences at the mRNA and protein levels, when placed in the context of the individual’s genetic background and exposure to environmental trauma, all determine the subtype and the phenotypic severity in each case. The translational implications of mutation analysis include improved diagnosis and subclassification, refined genetic counseling of families at risk, and development of DNA-based prenatal and preimplantation genetic diagnosis. The prospects of molecular therapies for epidermolysis bullosa include further development of strategies for gene therapy, protein replacement therapy and cell-based therapies, including stem cell therapy and bone marrow transfer. Collectively, advances in the molecular genetics of heritable skin diseases clearly emphasize the value of basic research for improved diagnostics and patient care for genetic skin diseases. Key words: heritable skin diseases; epidermolysis bullosa; prenatal testing; gene therapy.
(Accepted February 23, 2009.)
*This overview is based on Dr Uitto’s presentation at the International Symposium, “Progress in Dermatology”, in connection with the Uppsala University Hospital’s 300-year Anniversary, Sweden, on October 3, 2008.
Acta Derm Venereol 2009; 89: 228–235.
Jouni Uitto, Department of Dermatology and Cutaneous Biology, Jefferson Medical College, 233 S. 10th Street, Suite 450 BLSB, Philadelphia, PA 19107, USA. E-mail: Jouni.Uitto@jefferson.edu
CLINICAL SPECTRUM OF HERITABLE SKIN DISEASES
Heritable disorders with skin manifestations display a wide spectrum of phenotypic findings; at one end of the spectrum the clinical manifestations are relatively minor, limited to the skin, hair and nails, while at the other end of the spectrum the cutaneous manifestations are part of a multi-system pathology causing considerable morbidity and even mortality. Because most of the heritable skin diseases are relatively rare, many of these conditions pose a clinical challenge to practitioners, who may not be aware of the salient clinical and genetic features. Furthermore, the complexity of classification schemes, frequently riddled with eponyms, has posed further challenges to timely and accurate diagnosis. In many of these conditions the histopathology is not diagnostic and the molecular basis remained unknown for a long period after initial description of the disease, thus precluding DNA-based diagnostic procedures. Over the past two decades, however, this situation has changed drastically, and with the advent of technologies and strategies of molecular biology in general an increasing number of gene defects has been identified in specific genodermatoses (1). In fact, mutations are now known to occur in close to 300 distinct genes in such a manner that these genetic lesions explain the phenotypic manifestations characteristic of a heritable skin disease.
PREDICTABLE AND SURPRISING CANDIDATE GENES
Examination of the existing mutation databases in these diseases has revealed a number of genes that probably could have been predicted early on, on the basis of clinical, histopathologic, immunohistochemical, and/or ultrastructural findings, to serve as candidate gene/pro-tein systems. An example of such predictable genes is the group of skin fragility syndromes, epidermolysis bullosa (EB), in which we postulated initially that mutations in the structural genes expressed within the cutaneous basement membrane zone (BMZ) could explain the clinical phenotypes (2). This postulate has now been verified by demonstration of a large number of distinct mutations in as many as ten different genes expressed within the cutaneous BMZ (see below). At the same time, a number of mutated genes has turned out to be rather surprising, and the exact relationship of the mutations in the affected genes with the clinical and morphologic features in the patients’ skin is, in some cases, not well understood. For example, pseudoxanthoma elasticum (PXE), which manifests clinically primarily in the skin, the eyes, and the cardiovascular system, is now known to be caused by mutations in the ABCC6 gene, which is expressed primarily in the liver and the kidneys (3). More specifically, the precise function of the protein encoded by the ABCC6 gene, and the pathomechanistic links between the underlying genetic mutations and ectopic mineralization of connective tissue in the skin, the eyes, and the arterial blood vessels remain unclear.
To highlight the progress made in understanding the genetic basis of heritable skin diseases in general, this overview will summarize the progress made in molecular diagnostics in EB, review the clinical implications of the mutation analysis, and evaluate the prospects of molecular therapies for the treatment of this, currently intractable, group of blistering disorders.
CLINICAL AND GENETIC FEATURES OF EPIDERMOLYSIS BULLOSA
The heritable forms of EB consist of a group of mechano-bullous disorders, with skin fragility and blistering as the unifying diagnostic feature (4, 5). There is no ethnic or racial predilection, and EB has been encountered globally in different ancestral backgrounds. Although EB is considered to be an “orphan disease” (with fewer than 200,000 affected individuals in the USA and fewer than 1 in 2000 citizens in the European Union), there may be as many as 30,000–40,000 affected individuals in the USA, and close to 500,000 patients with EB worldwide.
The key clinical observation in EB is that the severity of skin manifestations is highly variable, reflecting in part the level of tissue separation within the skin (Table I). At one end of the spectrum, EB can manifest with relatively minor fragility as a result of trauma to the hands and feet, with minimal effects on the affected individual’s longevity. At the other end of the spectrum, skin fragility can lead to early demise of the affected individual within a few days or weeks of birth. Adding to the phenotypic complexity is the finding of extracutaneous manifestations that can be encountered in different subtypes of EB (4). Historically, this complexity, when coupled with different eponyms, has lead to suggestions that there are as many as 30 different subtypes of EB. Traditionally, however, EB has been divided into three broad categories based on the level of tissue separation within the cutaneous BMZ, as visualized by diagnostic electron microscopy or by immunoepitope mapping (Fig. 1 and Table I). In the classic simplex forms, tissue separation occurs within the basal keratinocytes, which lyse as a result of minor trauma. In the classic junctional forms of EB, tissue separation occurs within the lamina lucida of the cutaneous basement membrane, and in dystrophic, the severely scarring forms of EB, tissue separation occurs below the lamina densa, within the upper papillary dermis at the level of anchoring fibrils (4). In addition, we have suggested an additional category, the hemidesmosomal variant, which shows tissue cleavage at the basal cell plasma membrane/lamina lucida interface (6). While recognition of this category has been extremely helpful in identifying candidate genes in EB, the latest consensus classification (4), which is based on combinations of clinical and non-molecular laboratory findings on the level of tissue cleavage within the skin, does not recognize this as a distinct entity.
Table I. Molecular classification of epidermolysis bullosa (EB) with associated mutant genesa
| EB subtype | Inheritance | Level of tissue separation | Genesb |
| Simplex | AD, AR | Basal cells | KRT5, KRT14 |
| Hemidesmosomal GABEB EB-PA EB-MD | AR | Hemidesmosomes (both intra- and extracellular) | COL17A1, LAMB3 ITGB4, PLEC1, ITGA6 PLEC1 |
| Junctional | AR | Lamina lucida | LAMA3, LAMB3, LAMC2, COL17A1 |
| Dystrophic | AD, AR | Sub-lamina densa | COL7A1 |
aDifferent mutations within each gene have been noted to result in a spectrum of phenotypes (see text); for this reason, multiple genes are listed under each subtype.
bFor detailed genotype-phenotype correlations, see refs 5–8.
GABEB: generalized atrophic benign EB; EB-PA: EB with pyloric atresia; EB-MD: EB with muscular dystrophy; AD: autosomal dominant; AR: autosomal recessive.
Fig. 1. Complexity of the cutaneous basement membrane zone and molecular-based classification of epidermolysis bullosa. The figure schematically depicts basal keratinocytes at the lower part of the epidermis, separated from the papillary dermis by a dermal-epidermal basement membrane. Ultrastructurally recognizable attachment complexes and structural components of the basement membrane zone are indicated on the left, while specific proteins localized within each layer are indicated on the right. The level of tissue separation within each subgroup of epidermolysis bullosa is shown on the right. (Adapted from ref. 7, with permission).
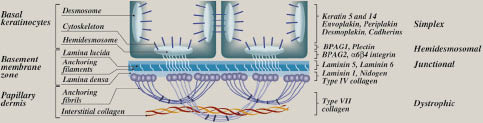
Critical for understanding the pathophysiology of EB is the ultrastructural recognition of distinct attachment complexes within the cutaneous BMZ; these include hemidesmosomes, which extend from the intracellular milieu of basal keratinocytes to the extracellular space, anchoring filaments that traverse the lamina lucida, and the anchoring fibrils, which extend from the lower part of the cutaneous basement membrane to the underlying dermis (Fig. 1). These structures form a contiguous network necessary for stabilization of the basal keratinocytes to the underlying basement membrane and its attachment to the papillary dermis. Thus, genetic alterations, such as premature termination codon mutations that result in synthesis of a truncated protein, or more discrete missense mutations that change the critical protein/protein interactions, can result in weakness of this network structure and manifest clinically as skin fragility and blistering characteristic of EB (5).
GENETIC BASIS OF EPIDERMOLYSIS BULLOSA
EB is inherited either in an autosomal dominant or autosomal recessive mode. Underlying mutations have now been identified in ten structural genes expressed within the cutaneous BMZ, and these mutations in these genes account for essentially all major forms of EB (4, 7, 8). It should be noted that, in some cases, mutations in the same gene can cause either autosomal dominant or autosomal recessive form of the disease, as exemplified by mutations in the type VII collagen gene (COL7A1), resulting either in dominantly or recessively inherited dominant dystrophic EB (DDEB and RDEB, respectively) (8). Furthermore, comparison of the mutation database with the clinical features in the affected individuals has allowed us to establish general genotype/phenotype correlations (5). Specifically, the level of expression of the mutated genes within the cutaneous BMZ, the types and combinations of mutations and their consequences at the mRNA and protein levels, when placed in the context of the individual’s genetic background and exposure to environmental trauma, determine the general subtype and the severity of the phenotype in each case. It should be noted that the latest report on classification of inherited EB (4) suggests inclusion of additional entities with distinct mutated genes. For example, lethal acantholytic EB (9) and plakophilin-deficient skin fragility/ectodermal dysplasia syndrome (10–12) have been recommended for addition in the EB simplex category, with the suprabasal level of cleavage. These entities harbor mutations in the desmoplakin (DSP) and plakophilin-1 (PKP-1) genes, respectively. Furthermore, Kindler syndrome, with mixed level of skin cleavage, has been suggested to be a distinct variant of EB. This disease is due to mutations in the kindlin-1 gene (KIND-1) encoding an actin-associated, keratinocyte focal contact point protein (13, 14). Finally, a form of EB simplex with migratory circinate erythema has been shown to result from a frameshift and delayed termination codon mutation in the keratin 5 gene (15).
A large number of mutations in the ten genes underlying the major forms of EB has been encountered over the past two decades, and for example, the Debra Molecular Diagnostics Laboratory, which was established in 1996 in the Department of Dermatology and Cutaneous Biology at Jefferson Medical College, has analyzed (as of 1 January 2009) 1012 families with EB for mutations (7, 8). A total of 781 distinct mutations where found in these ten different genes, indicating that there are relatively few recurrent mutations in EB and that most families harbor “private”, family specific mutations. Thus, even with streamlined mutation detection strategies, a considerable amount of work is involved in identifying the specific mutations in new families with EB.
TRANSLATIONAL IMPLICATIONS OF MUTATION ANALYSIS
Besides providing information on the specific mutation(s) in each affected individual, a legitimate question relates to the implications of the molecular diagnostics in EB. In other words, how does information on mutations benefit patients and their parents, and how does such information assist healthcare providers in delivering optimal care? It is very clear that molecular diagnostics on EB, with establishment of genotype/phenotype correlations, has improved the accuracy of diagnosis and subclassification with prognostic implications. The identification of mutations also has profound consequences for genetic counseling, particularly in families with no previous history of EB. For example, identification of specific type VII collagen mutations in a newborn with no family history and with relatively mild clinical features is able to determine whether the patient has a mild recessive dystrophic EB or whether the disease is a result of a de novo dominant mutation (16). Finally, a profound translational impact of the mutation analysis relates to the development of DNA-based prenatal testing in families at risk for recurrence (17). This can now be performed from chorionic villus sampling (CVS) as early as the tenth week of gestation, thus providing the parents and healthcare providers with critical information on the fetal genotype during the first trimester. It should be noted that DNA-based prenatal testing has essentially completely replaced the previously employed fetal skin biopsy that usually takes place between the 18th and the 20th week gestation.
FUTURE IMPACT OF MOLECULAR GENETICS ON EPIDERMOLYSIS BULLOSA
Preimplantation genetic diagnosis
As an extension of DNA-based prenatal testing, the feasibility of preimplantation genetic diagnosis for EB and related blistering disorders has already been established (18, 19). In this procedure, DNA-based diagnosis is performed even before the pregnancy is initiated in the context of in vitro fertilization. Specifically, blastomer analysis of eight-cell embryos can be performed and only the embryos that lack the deleterious mutations are implanted to the uterus for initiation of the pregnancy. The concept of preimplantation genetic diagnosis can be attractive in the sense that it completely avoids the consideration of termination of pregnancy. However, this procedure, while technically feasible, has not gained wide acceptance for EB, in part due to cost and to the elaborate clinical involvement of the mother.
Fetal cells and free DNA in maternal blood
Another intriguing development of DNA-based prenatal diagnosis entails examination of fetal cells or free fetal DNA in maternal blood (see Fig. 2) (20–23). The potential benefits of non-invasive prenatal diagnosis from maternal circulation would include avoidance of complications of invasive procedures, such as amniocentesis or CVS, which include small, yet clearly increased, risk for fetal loss. It is also hoped that these procedures would provide information earlier than currently allowed by CVS based testing, which can be performed as early as the 10th week of gestation. In fact, analysis of fetal DNA in the mother’s blood may provide information of the fetal genome as early as the fifth week of gestation, i.e. just at the time a woman becomes aware of her pregnancy (Fig. 3).
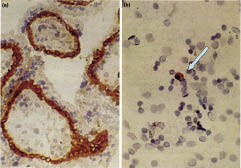
Fig. 2. Identification of fetal cytotrophoblasts in maternal blood using a monoclonal antibody (mAbMG2). (a) Immunohistochemical staining of first trimester placenta reveals strong expression of an intracellular antigen (red color). (b) An MG2-positive cell was also identified in maternal blood (arrow). (Adapted from ref. 21, with permission).
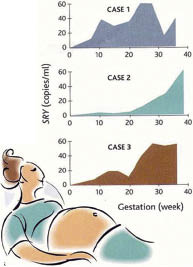
Fig. 3. Detection of free fetal DNA in the peripheral blood of three pregnant women (cases 1–3), as detected by quantitative PCR of sex determining region Y (SRY) representing DNA from male fetus. Note that clearly detectable levels of SRY are present in some cases as early as at the 5th week gestation. (Adapted from ref. 26, with permission).
Fetal cells are clearly present in the maternal circulation, but the estimates of the exact number, mostly based on polymerase chain reaction (PCR) amplification, vary widely (21). However, one estimate, based on Y chromosome-specific probe, identified 2–6 fetal cells per ml of maternal blood in mid-gestation (24). Also, a number of cell types, such as trophoblasts, mesenchymal stem cells, nucleated erythrocytes (CD71+), and hematopoietic progenitor cells (CD34+), have been identified and enriched by a number of enrichment and/or isolation techniques, including flow cytometry, magnetic bead separation, density gradient centrification, and laser capture microscopy (20). There are, however, critical questions relating to the use of fetal cells in maternal peripheral blood for prenatal testing: How early are they detectable and how long do they persist in circulation after pregnancy? In fact, some cell types have been suggested to persist in maternal circulation for years after the pregnancy, thus complicating the analysis in subsequent pregnancies (25). Also, the isolation procedures have to be optimized and the reliability of single cell PCR verified before such analysis can be attempted. Nevertheless, successful prenatal diagnosis utilizing fetal cells in the maternal peripheral blood as the source of DNA has already been performed in case of β-thalassemia, sickle-cell anemia, lamellar ichthyosis, and other conditions (see 20, 21).
Another intriguing observation is that free fetal DNA is easily detectable in the maternal circulation by PCR, and its concentration increases progressively during the pregnancy. In fact ∼3–6% of the total free DNA in the maternal circulation at term is of fetal origin (Fig. 3) (23, 26). Unlike fetal cells, the fetal DNA is cleared very rapidly after delivery, no trace being found after 24–48 h (27). The free fetal DNA probably originates from trophoblast breakdown, as free fetal RNA, which is placentally derived (choriogonadotropin and placental lactogen), can also been detected in the maternal circulation (28). An obvious problem in analyzing fetal DNA in the mother’s blood is that the maternal allele in fetal DNA is masked by maternal DNA. Consequently, this approach allows reliable screening only for the paternal allele in case of dominant mutations or in compound heterozygotes with different paternal and maternal mutations. Nevertheless, this technique has been used successfully for prenatal diagnosis for a number of heritable conditions, and it has been applied to analysis of the fetal RHD gene locus in RHD-negative mothers. In fact, several countries within the European community are in the process of, or are contemplating, implementing RHD genotyping of all fetuses of D-negative women by non-invasive prenatal diagnosis (29). It is conceivable, therefore, that these approaches will provide new opportunities for prenatal diagnosis by pushing the diagnostic window to earlier stages of pregnancy using non-invasive approaches.
PROSPECTS FOR MOLECULAR THERAPIES
In spite of impressive progress in molecular diagnostics of EB, no specific or effective treatment is currently available, short of control of infections and prevention of trauma. The progress in understanding the molecular basis of EB has suggested, however, that this condition is an excellent candidate disease for molecular correction strategies, including gene therapy, protein replacement therapy and cell-based therapies (30).
Gene therapy
In the context of gene therapy, specific molecular defects have been identified in ten different genes expressed in the cutaneous BMZ, and the majority of the mutations are single-base substitutions or small insertions or deletions, rather than complex chromosomal re-arrangements, and might therefore be more amenable to correction strategies. In addition, animal models that recapitulate the features of different subtypes of EB are readily available, including transgenic mice and naturally occurring mutations in cats, dogs, sheep, and large domestic animals (31). When contemplating gene therapy, the following general strategies need to be considered. (i) Should the procedures attempt gene replacement or repair of the defective gene? (ii) Should the correction take place ex vivo or in vivo? (iii) What is the optimal delivery system (viral vs. physical/chemical)? (iv) What is the appropriate preclinical model of the human disease?
One approach, an ex vivo gene therapy, contemplates the use of keratinocyte cultures established from the skin of an affected patient. These cells are then propagated and transduced with a cDNA-containing vector in culture, and the corrected keratinocytes are allowed to grow to epithelial sheets that are grafted back to the donor skin to a prepared acceptor site. The goals of the keratinocyte gene transfer are efficient targeting of stem cell populations, which would then allow durable expression of the transgene, leading to sustained synthesis of the protein and its assembly to supramolecular organization defective in the disease. The feasibility of this approach has already been demonstrated in the case of a junctional EB patient treated with a genetically modified keratinocyte grafts (32). An extended period of follow-up examination of the treated area (a total of ∼500 cm2) has revealed sustained phenotypic reversal and persistence of the skin graft. There has been no evidence of immune challenge to the graft or to the transduced cDNA product, the LAMB3 chain of laminin 332 (32). It should be noted, that this particular patient was carefully selected so that one of the underlying mutations (E210K) allowed low level of expression (< 5%) of this protein and, consequently, the patient’s immune system does not recognize the newly introduced polypeptide as a neoantigen. Similar experiments for additional patients with junctional forms of EB as well as for dystrophic (DEB) are currently contemplated, and it is hoped that the general applicability of this approach will become evident soon. The limitations of the DNA-based, vector-driven gene therapy include particular concerns of carcinogenesis due to integration of the constructs into the genome in a manner that would activate proto-oncogenes or inactivate tumor suppressor genes. In part due to these limitations and other potential drawbacks, complementary approaches utilizing protein replacement and cell transfer systems are being explored.
Protein replacement
The strategy of protein replacement therapy for genodermatoses will utilize recombinant proteins administered to the skin either topically, by local injection, or systemically via circulation. The goal is replacement of a defective protein or replenishment of a missing protein in a manner that would be amenable both for the autosomal dominant and autosomal recessive forms of the disease. The concept of protein therapeutics for EB was based on early observations that injection of recombinant human type VII collagen restores the collagen function in DEB in a xenograft model consisting of human EB skin on the back of immuno-compromised mice (33). More recently, injection of purified human recombinant type VII collagen into the circulation of mice with recessive DEB has been shown to result in incorporation of the injected collagen to the cutaneous BMZ, formation of anchoring fibrils, and amelioration of the blistering phenotype (34). In fact, the untreated mice usually die within the first week of life, while mice injected with the human type VII collagen survived up to 20–25 weeks of age (Fig. 4). It should be noted that the mice injected with human VII collagen developed antibodies recognizing this protein, but the antibodies did not recognize the mouse protein nor were they apparently pathogenic. Finally, the formation of antibodies recognizing human type VII collagen could be blocked by injection with an anti-CD40L monoclonal antibody (MR1) (34). Based on these studies, it is conceivable that protein replacement therapy may be an option in the future for treatment of recessive DEB, and similar work is currently underway for development of treatment for patients with junctional forms of EB due to mutations in the laminin 332 subunit polypeptide genes (35).
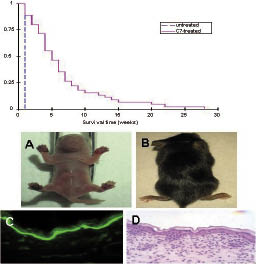
Fig. 4. Improved survival of recessive dystrophic epidermolysis bullosa mice (Col7a1–/–) injected with purified type VII collagen protein (C7). (Upper panel) Kaplan-Meier curves indicate that the untreated mice die within the first few days of life, while a significant increase in the survival, up to 20–25 weeks of life, of the mice treated with type VII collagen is noted (p < 0.001). (Lower panel) A 1-day old recessively inherited dominant dystrophic epidermolysis bullosa mouse depicts hemorrhagic blisters on paws (panel A); the same mouse survived past 22 weeks after injections with C7 (panel B). Immunofluorescence staining with an anti-type VII collagen antibody demonstrates signal at the cutaneous basement membrane zone of this 22-week old treated mouse (panel C), and histological analysis of the skin reveals apparently normal association of the epidermis and dermis (panel D). (Adapted from ref. 34, with permission).
Cell-based therapies
The latest attempts to develop molecular strategies for treatment of EB revolve around cell-based therapies. One set of studies has concentrated on human fibroblasts, which have been shown to home to skin wounds, deliver type VII collagen, and promote wound healing in RDEB mice when injected intravenously (36, 37). More recently, fibroblast therapy for RDEB has been tested by direct injection of the cells to the skin of the affected patients (38). In the latter study, injection of autologous and allogeneic fibroblasts increased the local type VII collagen expression in the RDEB patients’ skin, but the mechanism was determined to be primarily cytokine-mediated up-regulation of the mutant type VII collagen expression in those patients who had residual level of the synthetic activity from their mutant gene. Consequently, the patients who demonstrate some baseline type VII collagen synthesis seem to benefit from this approach, while those patients completely lacking type VII collagen expression might not (39).
More recently, the cell-based therapy for RDEB has been extended to utilize bone marrow stem cell transfer to RDEB mice (40, 41). Infusion of bone marrow cells, derived from green fluorescent protein (GFP) expressing mice, allowed tracing of the donor cells to the skin. In one study, a specific population of bone marrow derived cells, positive for the SLAM family receptor (CD150+/CD48–), were injected to RDEB mice and many of them survived for several months (41). The surviving animals had clear evidence of engraftment of donor cells in the skin, production of type VII collagen, and healing of skin blisters. In a similar approach, embryonic bone marrow cell transplantation has been shown to result in successful engraftment of the GFP-positive bone marrow cells in the skin, followed by differentiation towards fibroblast phenotypes and expression profiles, including deposition of type VII collagen primarily in the follicular BMZ (40). The embryonic bone marrow cell transfer also significantly ameliorated the severity of the dystrophic EB phenotype at birth, and the adult mice had an extended survival up to several weeks. An interesting observation in the mice subjected to embryonic bone marrow cell transfer was that these mice became tolerized to GFP, and subsequent grafting of GFP-containing skin did not induce the production of anti-GFP antibodies (Fig. 5) (40).
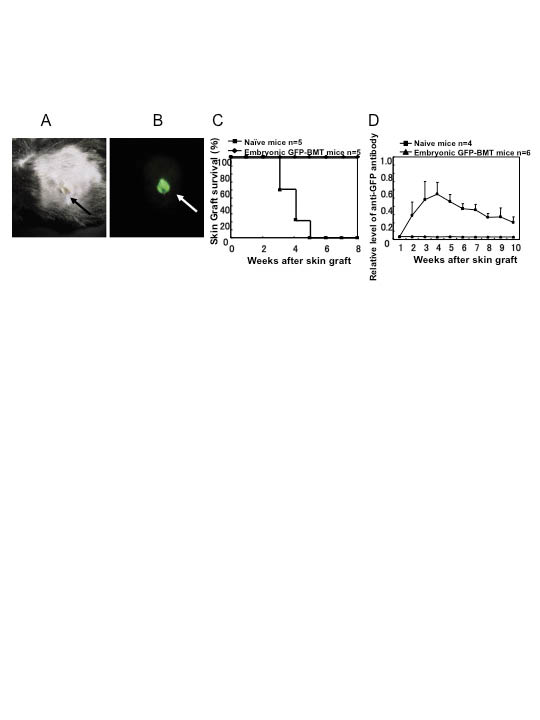
Fig. 5. Induction of tolerance to green fluorescent protein (GFP) by embryonic bone marrow transplant (BMT). (A, B) Skin from GFP-transgenic mouse tail is engrafted on a 7-week old mouse that had previously received embryonic GFP-bone marrow transfer. Engrafted GFP-transgenic skin can be observed under normal (A, black arrow) and fluorescent (B, white arrow) light. The engrafted GFP-transgenic skin persisted for more than 50 weeks. (C) Survival of GFP-tagged skin graft in mice treated with embryonic GFP-bone marrow transfer and in untreated, naïve mice. Engrafted skin persisted beyond 8 weeks after grafting in all mice that had undergone embryonic bone marrow transfer, while all skin grafts were rejected by non-irradiated, adult naïve mice before 5 weeks after grafting. (D) Generation of antibodies against GFP in transplanted skin measured using an enzyme-linked immunoassay (ELISA). Naïve mice (n = 4) produced significant amounts of antibodies (mean ± SE), whereas mice that had received embryonic GFP-bone marrow transfer (n = 6) did not. (Adapted from ref. 40, with permission).
Collectively, the preclinical observations utilizing an animal model for EB (Col7a1–/–) (42) provide “proof-of-principle” evidence in support of the possibility that application of allogeneic hematopoietic stem cell transplantation may be an option for treatment of human RDEB. In fact, based on these critical animal studies, a one-year old male with RDEB was recently infused with bone marrow cells derived from an HLA-matched older sibling donor. At 160 days after transplantation, type VII collagen and anchoring fibrils were documented in areas of healing skin by immunofluorescence and electron microscopy (Angela M. Christiano, personal communication). Additional patients are currently being treated with similar strategies, and the outcome of these interventions is being investigated. These early observations suggest that transfer of bone marrow cell populations can correct basement membrane defect in human RDEB and perhaps in other genetic skin diseases characterized by fibroblast dysfunction (40). However, refinement of strategies in experimental settings in specialized centers is required before these treatment modalities can be offered to the patient populations at large.
ACKNOWLEDGEMENTS
Carol Kelly assisted in preparation of this manuscript. Drs Mei Chen, Yasufumi Kaneda, Outi Kilpivaara, Katsuto Tamai and David Woodley kindly provided illustrations. The author’s original studies cited in this overview were supported by NIH/NIAMS grants P01AR38923 and R01AR55225, and by the Dermatology Foundation.
REFERENCES
